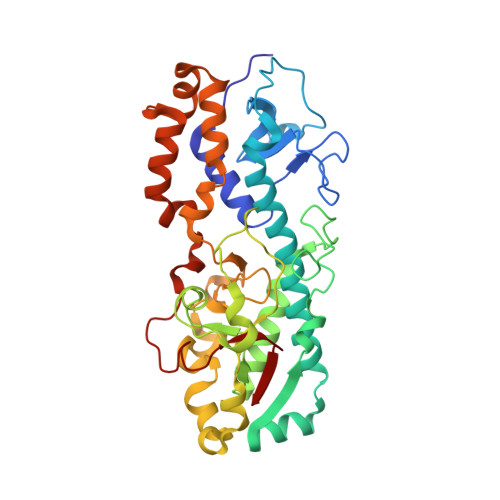
Structure of the reovirus outer capsid and dsRNA-binding protein sigma3 at 1.8 A resolution.
Olland, A.M., Jane-Valbuena, J., Schiff, L.A., Nibert, M.L., Harrison, S.C.(2001) EMBO J 20: 979-989
- PubMed: 11230122
- DOI: https://doi.org/10.1093/emboj/20.5.979
- Primary Citation of Related Structures:
1FN9 - PubMed Abstract:
The crystallographically determined structure of the reovirus outer capsid protein sigma3 reveals a two-lobed structure organized around a long central helix. The smaller of the two lobes includes a CCHC zinc-binding site. Residues that vary between strains and serotypes lie mainly on one surface of the protein; residues on the opposite surface are conserved. From a fit of this model to a reconstruction of the whole virion from electron cryomicroscopy, we propose that each sigma3 subunit is positioned with the small lobe anchoring it to the protein mu1 on the surface of the virion, and the large lobe, the site of initial cleavages during entry-related proteolytic disassembly, protruding outwards. The surface containing variable residues faces solvent. The crystallographic asymmetric unit contains two sigma3 subunits, tightly associated as a dimer. One broad surface of the dimer has a positively charged surface patch, which extends across the dyad. In infected cells, sigma3 binds dsRNA and inhibits the interferon response. The location and extent of the positively charged surface patch suggest that the dimer is the RNA-binding form of sigma3.
- Program in Virology, Division of Medical Sciences, Harvard Medical School, Harvard University, Cambridge, MA 02138, USA.
Organizational Affiliation: