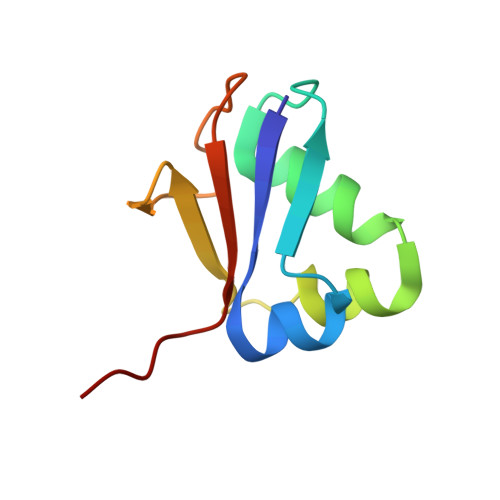
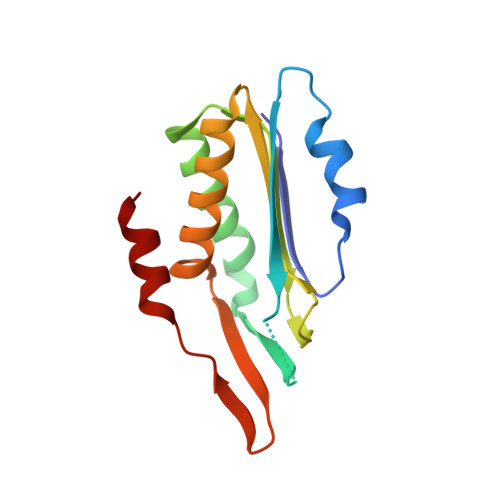
Crystal structure of molybdopterin synthase and its evolutionary relationship to ubiquitin activation.
Rudolph, M.J., Wuebbens, M.M., Rajagopalan, K.V., Schindelin, H.(2001) Nat Struct Biol 8: 42-46
- PubMed: 11135669
- DOI: https://doi.org/10.1038/83034
- Primary Citation of Related Structures:
1FM0, 1FMA - PubMed Abstract:
Molybdenum cofactor (Moco) biosynthesis is an evolutionarily conserved pathway present in eubacteria, archaea and eukaryotes, including humans. Genetic deficiencies of enzymes involved in Moco biosynthesis in humans lead to a severe and usually fatal disease. Moco contains a tricyclic pyranopterin, termed molybdopterin (MPT), that bears the cis-dithiolene group responsible for molybdenum ligation. The dithiolene group of MPT is generated by MPT synthase, which consists of a large and small subunits. The 1.45 A resolution crystal structure of MPT synthase reveals a heterotetrameric protein in which the C-terminus of each small subunit is inserted into a large subunit to form the active site. In the activated form of the enzyme this C-terminus is present as a thiocarboxylate. In the structure of a covalent complex of MPT synthase, an isopeptide bond is present between the C-terminus of the small subunit and a Lys side chain in the large subunit. The strong structural similarity between the small subunit of MPT synthase and ubiquitin provides evidence for the evolutionary antecedence of the Moco biosynthetic pathway to the ubiquitin dependent protein degradation pathway.
- Department of Biochemistry and Center for Structural Biology, State University of New York at Stony Brook, Stony Brook, New York 11794-5215, USA.
Organizational Affiliation: