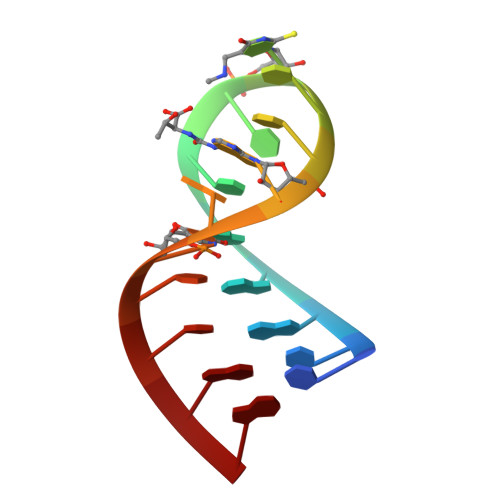
Hypermodified nucleosides in the anticodon of tRNALys stabilize a canonical U-turn structure.
Sundaram, M., Durant, P.C., Davis, D.R.(2000) Biochemistry 39: 12575-12584
- PubMed: 11027137
- DOI: https://doi.org/10.1021/bi0014655
- Primary Citation of Related Structures:
1FL8 - PubMed Abstract:
Modified nucleosides in the anticodon domain of Escherichia coli tRNA(Lys) are necessary for high-affinity codon recognition and reading frame maintenance. Human tRNA(Lys,3) is the specific primer for HIV-1 reverse transcriptase and also requires nucleoside modification for proper function. We now present NMR solution structures for the fully modified 17-nucleotide E. coli tRNA(Lys) anticodon stem-loop domain (ASL). NMR data were also collected for several partially modified ASLs, revealing the contributions each modified nucleoside (mnm(5)s(2)U34, t(6)A37, and psi39) makes in transforming the disordered, unmodified tRNA ASL into the highly ordered native structure. The solution structure of the native ASL domain provides insight into longstanding questions regarding both wobble position modification and the nearly ubiquitous t(6)A37 found in tRNAs with an adjacent U at position 36. Native tRNA(Lys) has a U-turn structure similar to the yeast tRNA(Phe) crystal structure, unlike previously proposed "unconventional" anticodon structures characterized by stable interactions between mnm(5)s(2)U-34 and t(6)A-37.
- Department of Medicinal Chemistry, University of Utah, Salt Lake City, Utah 84112, USA.
Organizational Affiliation: