The effect of intracellular molybdenum in Hydrogenophaga pseudoflava on the crystallographic structure of the seleno-molybdo-iron-sulfur flavoenzyme carbon monoxide dehydrogenase.
Hanzelmann, P., Dobbek, H., Gremer, L., Huber, R., Meyer, O.(2000) J Mol Biology 301: 1221-1235
- PubMed: 10966817
- DOI: https://doi.org/10.1006/jmbi.2000.4023
- Primary Citation of Related Structures:
1FFU, 1FFV - PubMed Abstract:
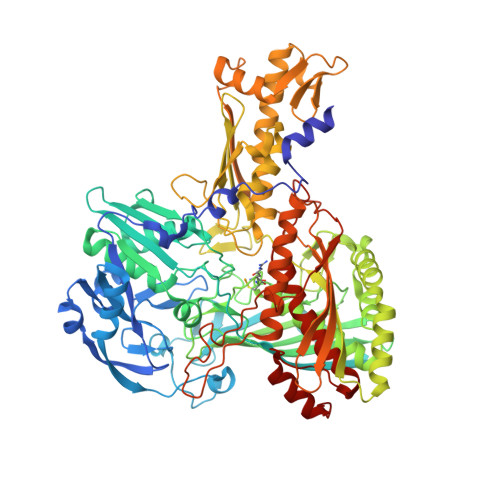
Crystal structures of carbon monoxide dehydrogenase (CODH), a seleno-molybdo-iron-sulfur flavoprotein from the aerobic carbon monoxide utilizing carboxidotrophic eubacterium Hydrogenophaga pseudoflava, have been determined from the enzyme synthesized at high (Mo(plus) CODH) and low intracellular molybdenum content (Mo(minus) CODH) at 2.25 A and 2.35 A resolution, respectively. The structures were solved by Patterson search methods utilizing the enzyme from Oligotropha carboxidovorans as the initial model. The CODHs from both sources are structurally very much conserved and show the same overall fold, architecture and arrangements of the molybdopterin-cytosine dinucleotide-type of molybdenum cofactor, the type I and type II [2Fe-2S] clusters and the flavin-adenine dinucleotide. Unlike the CODH from O. carboxidovorans, the enzyme from H. pseudoflava reveals a unique post-translationally modified C(gamma)-hydroxy-Arg384 residue which precedes the catalytically essential S-selanyl-Cys385 in the active-site loop. In addition, the Trp193 which shields the isoalloxazine ring of the flavin-adenine dinucleotide in the M subunit of the H. pseudoflava CODH is a Tyr193 in the O. carboxidovorans CODH. The hydrogen bonding interaction pattern of the molybdenum cofactor involves 27 hydrogen bonds with the surrounding protein. Of these, eight are with the cytosine moiety, eight with the pyrophosphate, six with the pyranopterin, and five with the ligands of the Mo ion. The structure of the catalytically inactive Mo(minus) CODH indicates that an intracellular Mo-deficiency affects exclusively the active site of the enzyme as an incomplete non-functional molybdenum cofactor was synthesized. The 5'-CDP residue was present in Mo(minus) CODH, whereas the Mo-pyranopterin moiety was absent. In Mo(plus) CODH the selenium faces the Mo ion and flips away from the Mo site in Mo(minus) CODH. The different side-chain conformations of the active-site residues S-selanyl-Cys385 and Glu757 in Mo(plus) and Mo(minus) CODH indicate a side-chain flexibility and a function of the Mo ion in the proper orientation of both residues.
- Lehrstuhl für Mikrobiologie, Universität Bayreuth, Universitätsstrasse 30, Bayreuth, D-95440, Germany.
Organizational Affiliation: