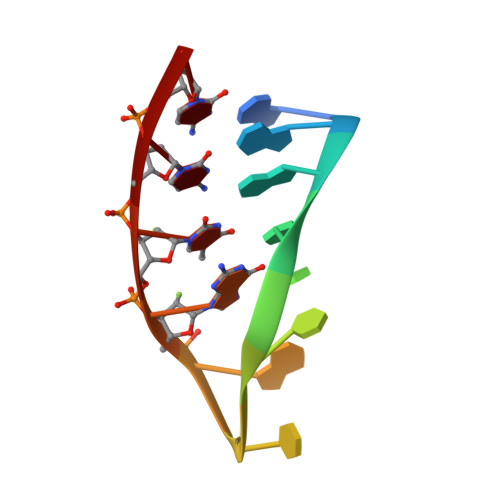
NMR solution structure of an oligonucleotide hairpin with a 2'F-ANA/RNA stem: implications for RNase H specificity toward DNA/RNA hybrid duplexes.
Trempe, J.F., Wilds, C.J., Denisov, A.Y., Pon, R.T., Damha, M.J., Gehring, K.(2001) J Am Chem Soc 123: 4896-4903
- PubMed: 11457316
- DOI: https://doi.org/10.1021/ja003859p
- Primary Citation of Related Structures:
1FC8 - PubMed Abstract:
The first structure of a 2'-deoxy-2'-fluoro-D-arabinose nucleic acid (2'F-ANA)/RNA duplex is presented. We report the structural characterization by NMR spectroscopy of a small hybrid hairpin, r(GGAC)d(TTCG)2'F-a(GTCC), containing a 2'F-ANA/RNA stem and a four-residue DNA loop. Complete (1)H, (13)C, (19)F, and (31)P resonance assignments, scalar coupling constants, and NOE constraints were obtained from homonuclear and heteronuclear 2D spectra. In the chimeric duplex, the RNA strand adopts a classic A-form structure having C3' endo sugar puckers. The 2'F-ANA strand is neither A-form nor B-form and contains O4' endo sugar puckers. This contrasts strongly with the dynamic sugar conformations previously observed in the DNA strands of DNA/RNA hybrid duplexes. Structural parameters for the duplex, such as minor groove width, x-displacement, and inclination, were intermediate between those of A-form and B-form duplexes and similar to those of DNA/RNA duplexes. These results rationalize the enhanced stability of 2'F-ANA/RNA duplexes and their ability to elicit RNase H activity. The results are relevant for the design of new antisense drugs based on sugar-modified nucleic acids.
- Department of Biochemistry and Montreal Joint Center for Structural Biology, McGill University, Montreal, Canada.
Organizational Affiliation: