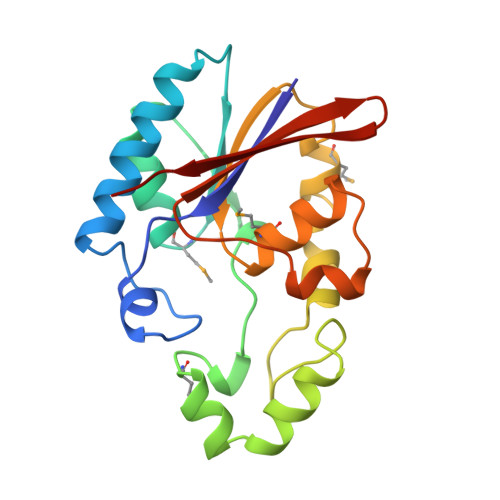
Crystal structure of the rat liver fructose-2,6-bisphosphatase based on selenomethionine multiwavelength anomalous dispersion phases.
Lee, Y.H., Ogata, C., Pflugrath, J.W., Levitt, D.G., Sarma, R., Banaszak, L.J., Pilkis, S.J.(1996) Biochemistry 35: 6010-6019
- PubMed: 8634242
- DOI: https://doi.org/10.1021/bi9600613
- Primary Citation of Related Structures:
1FBT - PubMed Abstract:
The crystal structure of the recombinant fructose-2,6-bisphosphatase domain, which covers the residues between 251 and 440 of the rat liver bifunctional enzyme, 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase, was determined by multiwavelength anomalous dispersion phasing and refined at 2.5 A resolution. The selenomethionine-substituted protein was induced in the methionine auxotroph, Escherichia coli DL41DE3, purified, and crystallized in a manner similar to that of the native protein. Phase information was calculated using the multiwavelength anomalous dispersion data collected at the X-ray wavelengths near the absorption edge of the K-shell alpha electrons of selenium. The fructose-2,6-bisphosphatase domain has a core alpha/beta structure which consists of six stacked beta-strands, four parallel and two antiparallel. The core beta-sheet is surrounded by nine alpha-helices. The catalytic site, as defined by a bound phosphate ion, is positioned near the C-terminal end of the beta-sheet and close to the N-terminal end of an alpha-helix. The active site pocket is funnel-shaped. The narrow opening of the funnel is wide enough for a water molecule to pass. The key catalytic residues, including His7, His141, and Glu76, are near each other at the active site and probably function as general acids and/or bases during a catalytic cycle. The inorganic phosphate molecule is bound to an anion trap formed by Arg6, His7, Arg56, and His141. The core structure of the Fru-2,6-P2ase is similar to that of the yeast phosphoglycerate mutase and the rat prostatic acid phosphatase. However, the structure of one of the loops near the active site is completely different from the other family members, perhaps reflecting functional differences and the nanomolar range affinity of Fru-2,6-P2ase for its substrate. The imidazole rings of the two key catalytic residues, His7 and His141, are not parallel as in the yeast phosphoglycerate mutase. The crystal structure is used to interpret the existing chemical data already available for the bisphosphatase domain. In addition, the crystal structure is compared with two other proteins that belong to the histidine phosphatase family.
- Department of Biochemistry, University of Minnesota, Minneapolis 55455, USA.
Organizational Affiliation: