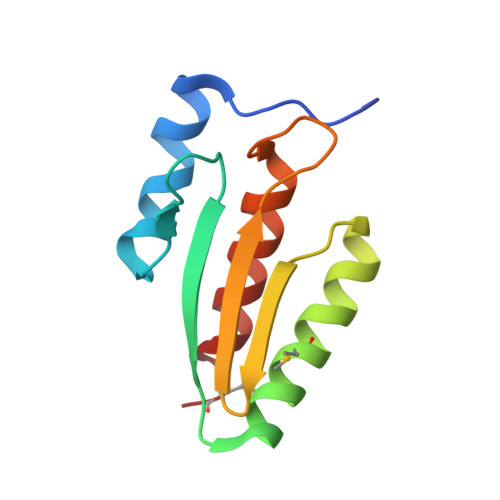
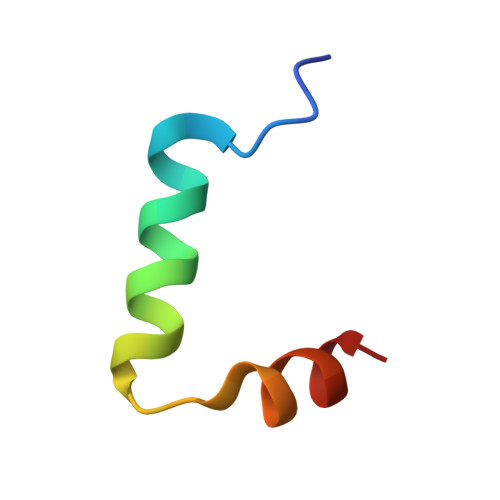
Structural basis of dimerization, coactivator recognition and MODY3 mutations in HNF-1alpha.
Rose, R.B., Bayle, J.H., Endrizzi, J.A., Cronk, J.D., Crabtree, G.R., Alber, T.(2000) Nat Struct Biol 7: 744-748
- PubMed: 10966642
- DOI: https://doi.org/10.1038/78966
- Primary Citation of Related Structures:
1F93 - PubMed Abstract:
Maturity-onset diabetes of the young type 3 (MODY3) results from mutations in the transcriptional activator hepatocyte nuclear factor-1alpha (HNF-1alpha). Several MODY3 mutations target the HNF-1alpha dimerization domain (HNF-p1), which binds the coactivator, dimerization cofactor of HNF-1 (DCoH). To define the mechanism of coactivator recognition and the basis for the MODY3 phenotype, we determined the cocrystal structure of the DCoH-HNF-p1 complex and characterized biochemically the effects of MODY3 mutations in HNF-p1. The DCoH-HNF-p1 complex comprises a dimer of dimers in which HNF-p1 forms a unique four-helix bundle. Through rearrangements of interfacial side chains, a single, bifunctional interface in the DCoH dimer mediates both HNF-1alpha binding and formation of a competing, transcriptionally inactive DCoH homotetramer. Consistent with the structure, MODY3 mutations in HNF-p1 reduce activator function by two distinct mechanisms.
- Department of Molecular and Cell Biology, University of California, Berkeley, California 94720-3206, USA.
Organizational Affiliation: