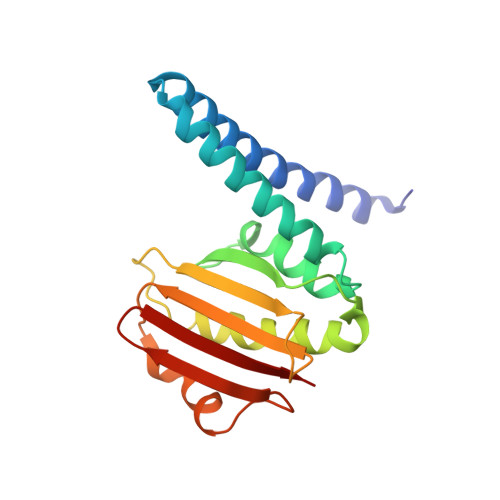
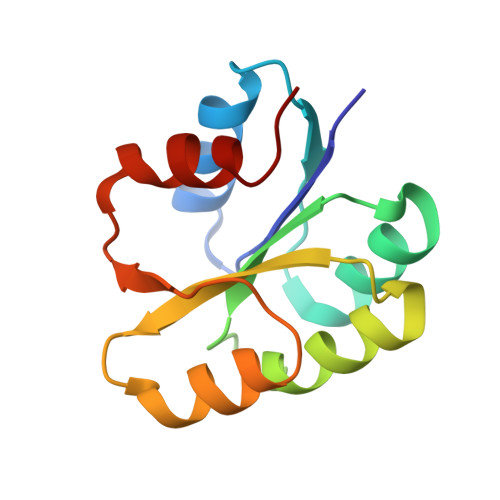
A transient interaction between two phosphorelay proteins trapped in a crystal lattice reveals the mechanism of molecular recognition and phosphotransfer in signal transduction.
Zapf, J., Sen, U., Madhusudan, M., Hoch, J.A., Varughese, K.I.(2000) Structure 8: 851-862
- PubMed: 10997904
- DOI: https://doi.org/10.1016/s0969-2126(00)00174-x
- Primary Citation of Related Structures:
1F51 - PubMed Abstract:
Spo0F and Spo0B specifically exchange a phosphoryl group in a central step of the phosphorelay signal transduction system that controls sporulation in Bacilli. Spo0F belongs to the superfamily of response regulator proteins and is one of 34 such proteins in Bacillus subtilis. Spo0B is structurally similar to the phosphohistidine domain of histidine kinases, such as EnvZ, and exchanges a phosphoryl group between His30 and Asp54 on Spo0F. Information at the molecular level on the interaction between response regulators and phosphohistidine domains is necessary to develop a rationale for how phospho-signaling fidelity is maintained in two-component systems. Structural analysis of a co-crystal of the Spo0F response regulator interacting with the Spo0B phosphotransferase of the phosphorelay signal transduction system of B. subtilis was carried out using X-ray crystallographic techniques. The association of the two molecules brings the catalytic residues from both proteins into precise alignment for phosphoryltransfer. Upon complex formation, the Spo0B conformation remains unchanged. Spo0F also retains the overall conformation; however, two loops around the active site show significant deviations. The Spo0F-Spo0B interaction appears to be a prototype for response regulator-histidine kinase interactions. The primary contact surface between these two proteins is formed by hydrophobic regions in both proteins. The Spo0F residues making up the hydrophobic patch are very similar in all response regulators suggesting that the binding is initiated through the same residues in all interacting response regulator-kinase pairs. The bulk of the interactions outside this patch are through nonconserved residues. Recognition specificity is proposed to arise from interactions of the nonconserved residues, especially the hypervariable residues of the beta4-alpha4 loop.
- Department of Molecular and Experimental Medicine, The Scripps Research Institute, La Jolla, CA 92037, USA.
Organizational Affiliation: