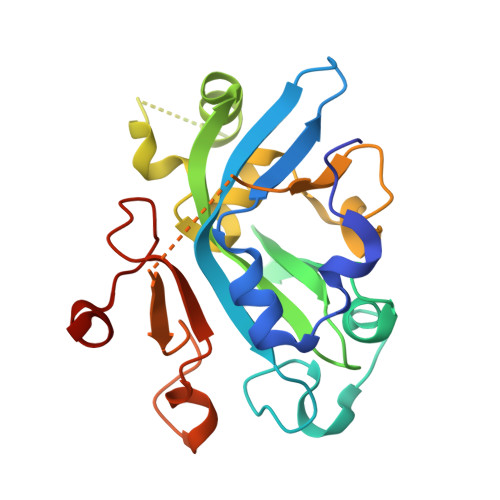
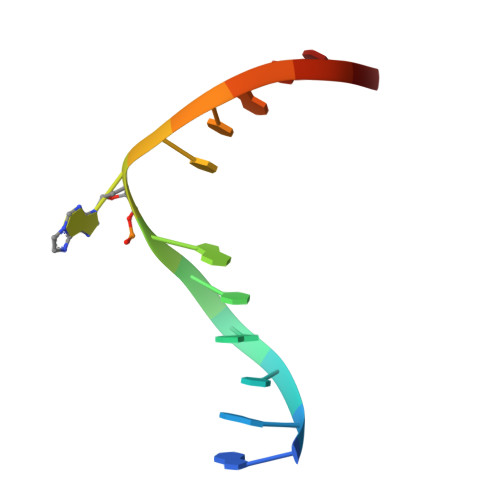
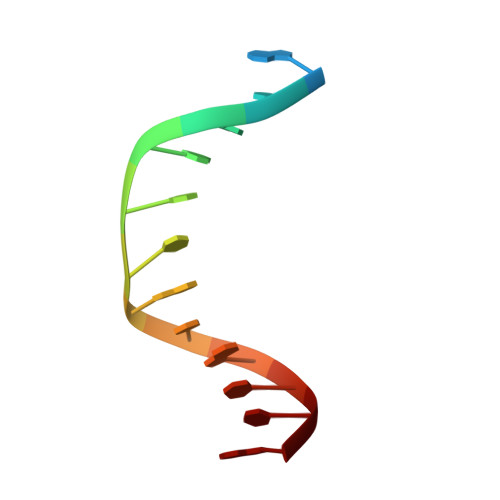
Molecular basis for discriminating between normal and damaged bases by the human alkyladenine glycosylase, AAG.
Lau, A.Y., Wyatt, M.D., Glassner, B.J., Samson, L.D., Ellenberger, T.(2000) Proc Natl Acad Sci U S A 97: 13573-13578
- PubMed: 11106395
- DOI: https://doi.org/10.1073/pnas.97.25.13573
- Primary Citation of Related Structures:
1EWN, 1F4R, 1F6O - PubMed Abstract:
The human 3-methyladenine DNA glycosylase [alkyladenine DNA glycosylase (AAG)] catalyzes the first step of base excision repair by cleaving damaged bases from DNA. Unlike other DNA glycosylases that are specific for a particular type of damaged base, AAG excises a chemically diverse selection of substrate bases damaged by alkylation or deamination. The 2.1-A crystal structure of AAG complexed to DNA containing 1,N(6)-ethenoadenine suggests how modified bases can be distinguished from normal DNA bases in the enzyme active site. Mutational analyses of residues contacting the alkylated base in the crystal structures suggest that the shape of the damaged base, its hydrogen-bonding characteristics, and its aromaticity all contribute to the selective recognition of damage by AAG.
- Department of Biological Chemistry and Molecular Pharmacology, Harvard Medical School, 240 Longwood Avenue, Boston, MA 02115, USA.
Organizational Affiliation: