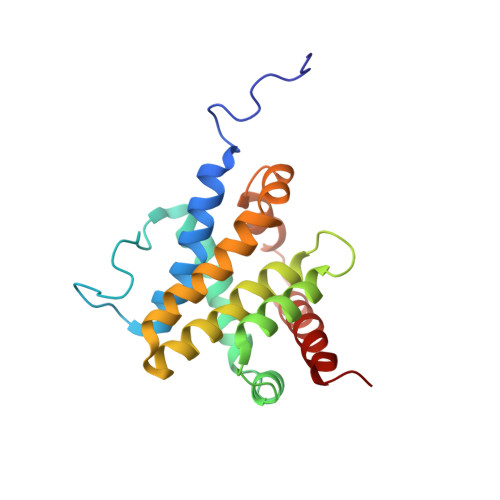
Structure of Bax: coregulation of dimer formation and intracellular localization.
Suzuki, M., Youle, R.J., Tjandra, N.(2000) Cell 103: 645-654
- PubMed: 11106734
- DOI: https://doi.org/10.1016/s0092-8674(00)00167-7
- Primary Citation of Related Structures:
1F16 - PubMed Abstract:
Apoptosis is stimulated by the insertion of Bax from the cytosol into mitochondrial membranes. The solution structure of Bax, including the putative transmembrane domain at the C terminus, was determined in order to understand the regulation of its subcellular location. Bax consists of 9 alpha helices where the assembly of helices alpha1 through alpha 8 resembles that of the apoptosis inhibitor, Bcl-x(L). The C-terminal alpha 9 helix occupies the hydrophobic pocket proposed previously to mediate heterodimer formation and bioactivity of opposing members of the Bcl-2 family. The Bax structure shows that the orientation of helix alpha 9 provides simultaneous control over its mitochondrial targeting and dimer formation.
- Biochemistry Section, Surgical Neurology Branch, National Institute of Neurological Disorders and Stroke, National Institutes of Health, Bethesda, MD 20892, USA.
Organizational Affiliation: