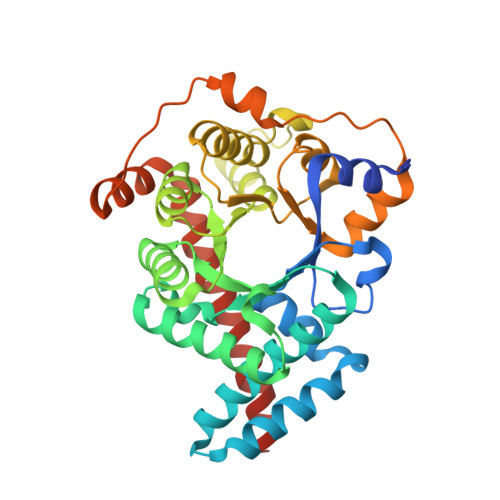
The three-dimensional structure of human transaldolase.
Thorell, S., Gergely Jr., P., Banki, K., Perl, A., Schneider, G.(2000) FEBS Lett 475: 205-208
- PubMed: 10869557
- DOI: https://doi.org/10.1016/s0014-5793(00)01658-6
- Primary Citation of Related Structures:
1F05 - PubMed Abstract:
The crystal structure of human transaldolase has been determined to 2.45 A resolution. The enzyme folds into an alpha/beta barrel structure and is thus similar in structure to other class I aldolases. Structure-based sequence alignment of available sequences of the transaldolase subfamily reveals that eight active site residues are invariant in the whole subfamily. Other invariant residues are mainly involved in the formation of the hydrophobic core of the enzyme. Noteworthy is a hydrophobic cluster consisting of five invariant residues. Human transaldolase has been implicated as an autoantigen in multiple sclerosis and four immunodominant peptide segments are located at the surface of the enzyme, accessible to autoantibodies.
- Department of Medical Biochemistry and Biophysics, Karolinska Institutet, Stockholm, Sweden.
Organizational Affiliation: