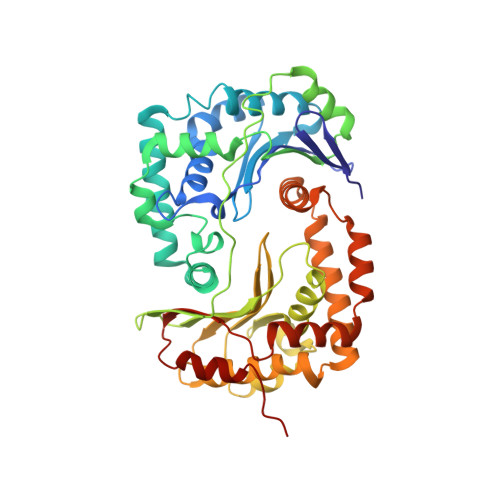
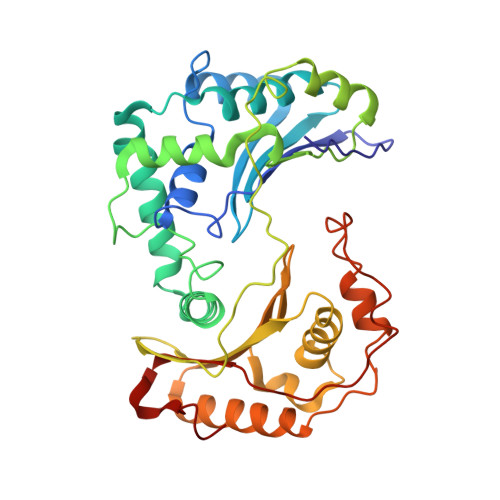
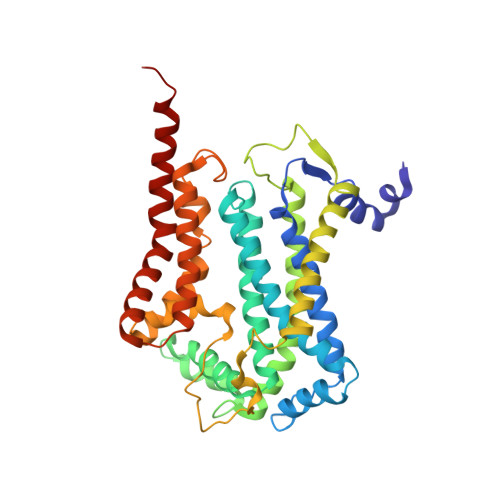
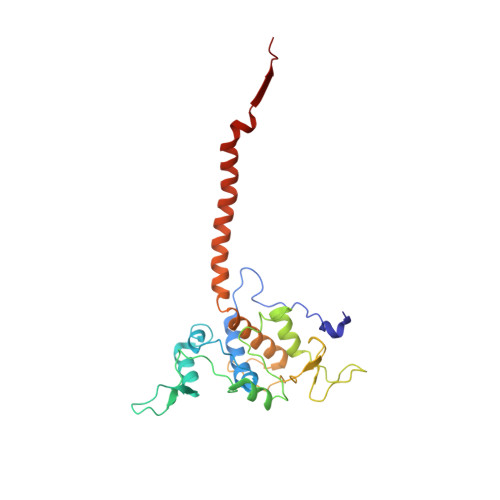
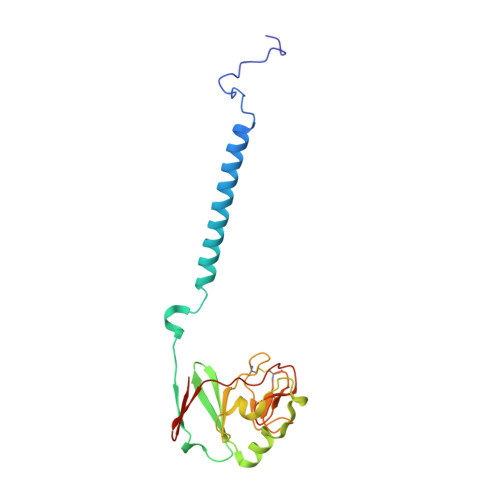
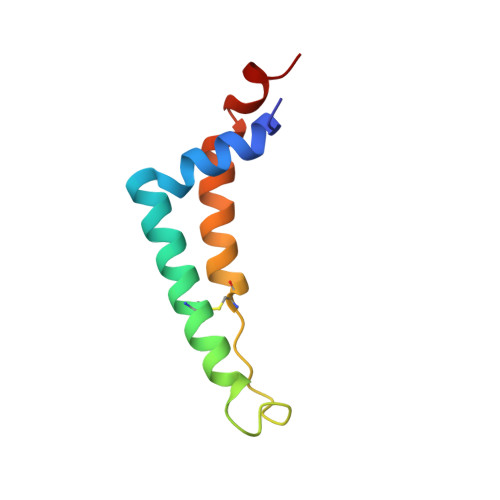
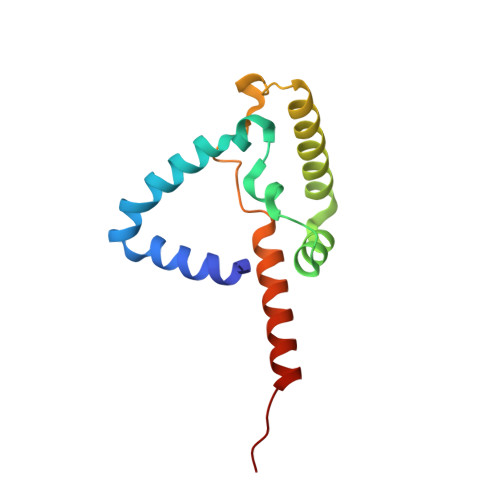
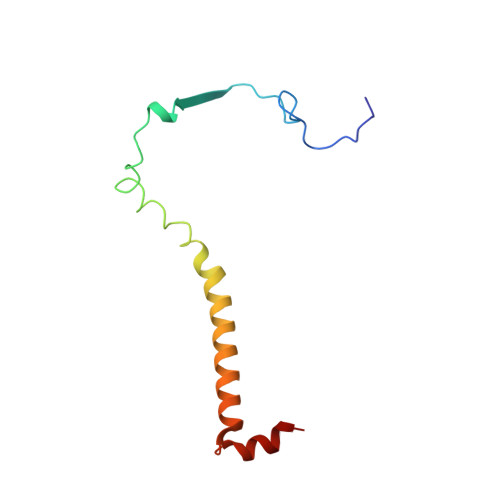
Structure at 2.3 A resolution of the cytochrome bc(1) complex from the yeast Saccharomyces cerevisiae co-crystallized with an antibody Fv fragment.
Hunte, C., Koepke, J., Lange, C., Rossmanith, T., Michel, H.(2000) Structure 8: 669-684
- PubMed: 10873857
- DOI: https://doi.org/10.1016/s0969-2126(00)00152-0
- Primary Citation of Related Structures:
1EZV - PubMed Abstract:
The cytochrome bc(1) complex is part of the energy conversion machinery of the respiratory and photosynthetic electron transfer chains. This integral membrane protein complex catalyzes electron transfer from ubiquinol to cytochrome c. It couples the electron transfer to the electrogenic translocation of protons across the membrane via a so-called Q cycle mechanism. The cytochrome bc(1) complex from the yeast Saccharomyces cerevisiae was crystallized together with a bound antibody Fv fragment. The structure was determined at 2.3 A resolution using multiple isomorphous replacement, and refined to a crystallographic R factor of 22.2% (R(free) = 25.4%). The complex is present as a homodimer. Each 'monomer' of the refined model includes 2178 amino acid residues of subunits COR1, QCR2, COB, CYT1, RIP1, QCR6, QCR7, QCR8 and QCR9 of the cytochrome bc(1) complex and of the polypeptides V(H) and V(L) of the Fv fragment, the cofactors heme b(H), heme b(L), heme c(1), the [2Fe-2S] cluster and 346 water molecules. The Fv fragment binds to the extrinsic domain of the [2Fe-2S] Rieske protein and is essential for formation of the crystal lattice. The approach to crystallize membrane proteins as complexes with specific antibody fragments appears to be of general importance. The structure of the yeast cytochrome bc(1) complex reveals in detail the binding sites of the natural substrate coenzyme Q6 and the inhibitor stigmatellin. Buried water molecules close to the binding sites suggest possible pathways for proton uptake and release. A comparison with other cytochrome bc(1) complexes shows features that are specific to yeast.
- Max-Planck-Institut für Biophysik, Abt. Molekulare Membranbiologie, Frankfurt, 60528, Germany. hunte@mpibp-frankfurt.mpg.de.
Organizational Affiliation: