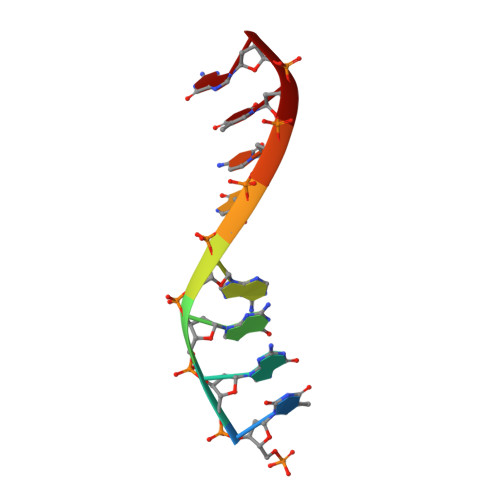
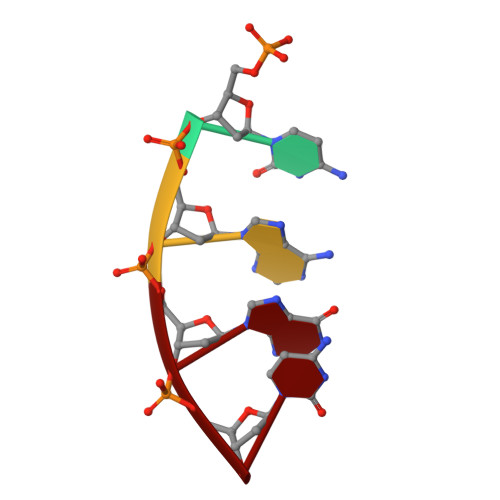
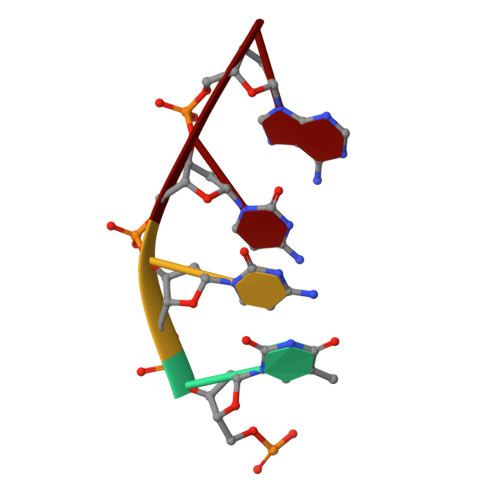
The NMR structure of estrone (Es)-tethered tandem DNA duplex: [d(5'pCAGCp3')-Es] + [Es-d(5'pTCCA3')]: d(5'pTGGAGCTG3').
Denisov, A.Y., Sandstrom, A., Maltseva, T.V., Pyshnyi, D.V., Ivanova, E.M., Zarytova, V.F., Chattopadhyaya, J.(1997) J Biomol Struct Dyn 15: 499-516
- PubMed: 9439997
- DOI: https://doi.org/10.1080/07391102.1997.10508961
- Primary Citation of Related Structures:
1ESS, 1TAN - PubMed Abstract:
The solution structure of an estrone (Es)-tethered tandem DNA duplex consisting of two Es-tethered tetranucleotides and a target octameric DNA sequence is reported. The structure of this Es-tethered tandem duplex has been compared with a corresponding natural tandem duplex without estrones. The Tm of the 3'-Es-tethered tetranucleotide part of the tandem duplex increases by 5 degrees C, whereas the Tm of the 5'-Es-tethered tetranucleotide part increases by 7 degrees C, compared with the corresponding natural counterpart. The NMR structures of both the Es-tethered tandem duplex and the natural counterpart have been based on 24 experimental NMR constraints per residue. Despite the fact that there is considerable distortion at the junction of two Es-tethered tetranucleotides in the major groove of the Es-tethered DNA duplex compared to the natural counterpart, both duplexes do take up B-type DNA structures. It is likely that the spatial proximity of two Es residues, and the resulting hydrophobic interaction between them might be responsible for the increase of the thermal stability of the Es-tethered tandem duplex in comparison with the natural counterpart.
- Department of Bioorganic Chemistry, University of Uppsala, Sweden.
Organizational Affiliation: