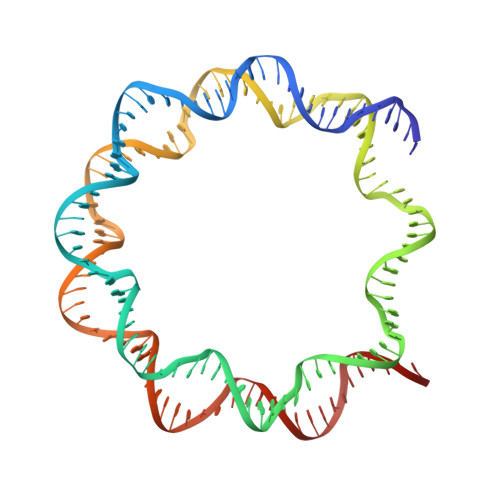
Asymmetries in the nucleosome core particle at 2.5 A resolution.
Harp, J.M., Hanson, B.L., Timm, D.E., Bunick, G.J.(2000) Acta Crystallogr D Biol Crystallogr 56: 1513-1534
- PubMed: 11092917
- DOI: https://doi.org/10.1107/s0907444900011847
- Primary Citation of Related Structures:
1EQZ - PubMed Abstract:
The 2.5 A X-ray crystal structure of the nucleosome core particle presented here provides significant additions to the understanding of the nucleosome, the fundamental unit of chromatin structure. Extensions are made to the structure of the N-terminal histone tails and details are provided on hydration and ion binding. The structure is composed of twofold symmetric molecules, native chicken histone octamer cores and the DNA palindrome, which were expected to form a perfectly twofold symmetric nucleosome core particle. In fact, the result is asymmetric owing to the binding of the DNA to the protein surface and to the packing of the particles in the crystal lattice. An analysis is made of the asymmetries by comparisons both within the nucleosome core particle and to the structure of the histone octamer core of the nucleosome.
- University of Tennessee/Oak Ridge Graduate Program for Genome Sciences and Technology, Oak Ridge National Laboratory, Oak Ridge, TN 37831-8080, USA.
Organizational Affiliation: