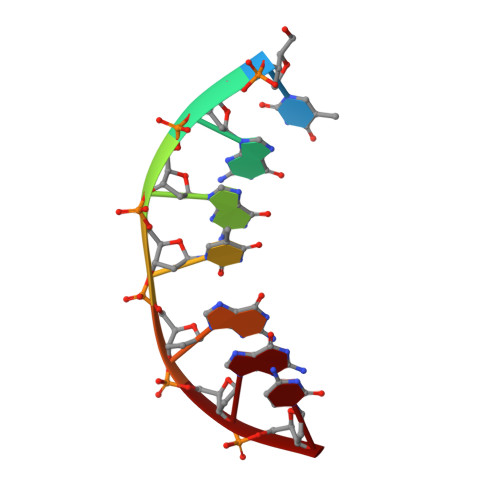
NMR observation of T-tetrads in a parallel stranded DNA quadruplex formed by Saccharomyces cerevisiae telomere repeats.
Patel, P.K., Hosur, R.V.(1999) Nucleic Acids Res 27: 2457-2464
- PubMed: 10352174
- DOI: https://doi.org/10.1093/nar/27.12.2457
- Primary Citation of Related Structures:
1EMQ - PubMed Abstract:
We report here the NMR structure of the DNA sequence d-TGGTGGC containing two repeats of Saccharomyces cerevisiae telomere DNA which is unique in that it has a single thymine in the repeat sequence and the number of Gs can vary from one to three. The structure is a novel quadruplex incor-porating T-tetrads formed by symmetrical pairing of four Ts via O4-H3 H-bonds in a plane. This is in contrast to the previous results on other telomeric sequences which contained more than one T in the repeat sequences and they were seen mostly in the flexible regions of the structures. We observed that the T4-tetrad was nicely accommodated in the center of the G-quadruplex, but it caused a small underwinding of the right handed helix. The T tetrad stacked well on the adjacent G3-tetrad, but poorly on the G5 tetrad. Likewise, T1 also formed a stable T-tetrad at the 5' end of the quadruplex. To our knowledge, this is the first report of T-tetrad formation in DNA structures. These observations are of significance from the points of view of both structural diversity and specific recognitions.
- Department of Chemical Sciences, Tata Institute of Fundamental Research, Homi Bhabha Road, Mumbai 400 005, India.
Organizational Affiliation: