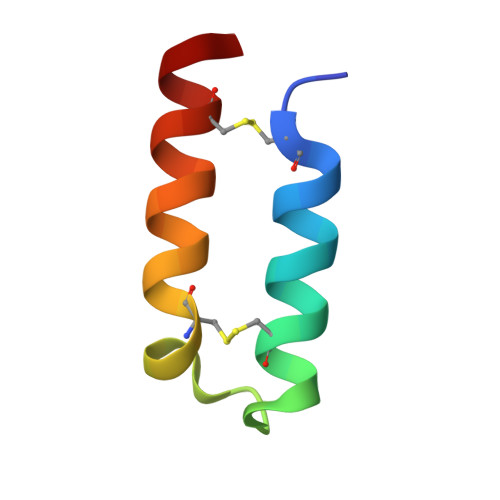
Synthesis and NMR solution structure of an alpha-helical hairpin stapled with two disulfide bridges.
Barthe, P., Rochette, S., Vita, C., Roumestand, C.(2000) Protein Sci 9: 942-955
- PubMed: 10850804
- DOI: https://doi.org/10.1110/ps.9.5.942
- Primary Citation of Related Structures:
1EI0 - PubMed Abstract:
Helical coiled-coils and bundles are some of the most common structural motifs found in proteins. Design and synthesis of alpha-helical motifs may provide interesting scaffolds that can be useful as host structures to display functional sites, thus allowing the engineering of novel functional miniproteins. We have synthesized a 38-amino acid peptide, alpha2p8, encompassing the alpha-helical hairpin present in the structure of p8MTCP1, as an alpha-helical scaffold particularly promising for its stability and permissiveness of sequence mutations. The three-dimensional structure of this peptide has been solved using homonuclear two-dimensional NMR techniques at 600 MHz. After sequence specific assignment, a total of 285 distance and 29 dihedral restraints were collected. The solution structure of alpha2p8 is presented as a set of 30 DIANA structures, further refined by restrained molecular dynamics, using simulated annealing protocol with the AMBER force field. The RMSD values for the backbone and all heavy atoms are 0.65+/-0.25 and 1.51+/-0.21 A, respectively. Excised from its protein context, the alpha-hairpin keeps its native structure: an alpha-helical coiled-coil, similar to that found in superhelical structures, with two helices spanning residues 4-16 and 25-36, and linked by a short loop. This motif is stabilized by two interhelical disulfide bridges and several hydrophobic interactions at the helix interface, leaving most of its solvent-exposed surface available for mutation. This alpha-helical hairpin, easily amenable to synthetic chemistry and biological expression system, may represent a stable and versatile scaffold to display new functional sites and peptide libraries.
- Centre de Biochimie Structurale, CNRS-UMR 9955, INSERM-U414, Université de Montpellier I, Faculté de Pharmacie, France.
Organizational Affiliation: