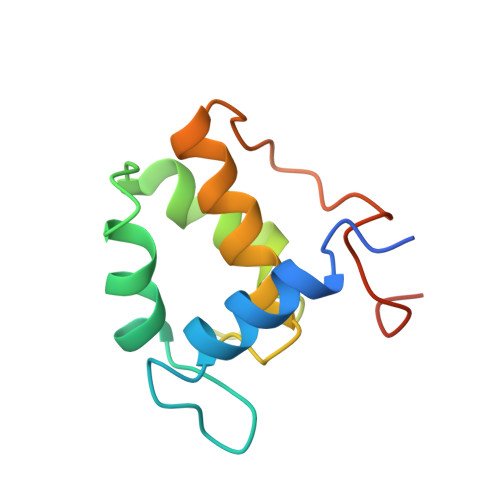
Structure and Asn-Pro-Phe binding pocket of the Eps15 homology domain.
de Beer, T., Carter, R.E., Lobel-Rice, K.E., Sorkin, A., Overduin, M.(1998) Science 281: 1357-1360
- PubMed: 9721102
- DOI: https://doi.org/10.1126/science.281.5381.1357
- Primary Citation of Related Structures:
1EH2 - PubMed Abstract:
Eps15 homology (EH) domains are eukaryotic signaling modules that recognize proteins containing Asn-Pro-Phe (NPF) sequences. The structure of the central EH domain of Eps15 has been solved by heteronuclear magnetic resonance spectroscopy. The fold consists of a pair of EF hand motifs, the second of which binds tightly to calcium. The NPF peptide is bound in a hydrophobic pocket between two alpha helices, and binding is mediated by a critical aromatic interaction as revealed by structure-based mutagenesis. The fold is predicted to be highly conserved among 30 identified EH domains and provides a structural basis for defining EH-mediated events in protein trafficking and growth factor signaling.
- Department of Pharmacology, University of Colorado Health Sciences Center, 4200 East Ninth Avenue, Denver, CO 80262, USA.
Organizational Affiliation: