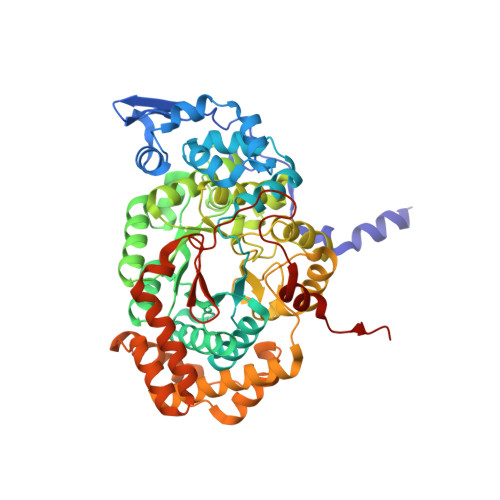
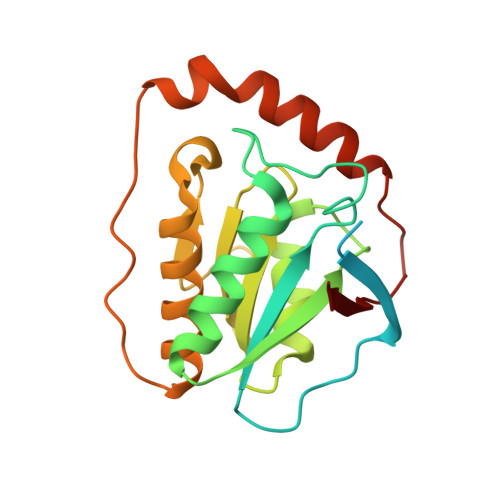
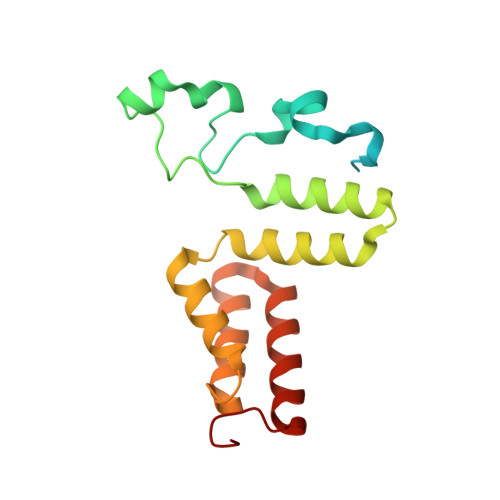
How a protein generates a catalytic radical from coenzyme B(12): X-ray structure of a diol-dehydratase-adeninylpentylcobalamin complex.
Masuda, J., Shibata, N., Morimoto, Y., Toraya, T., Yasuoka, N.(2000) Structure 8: 775-788
- PubMed: 10903944
- DOI: https://doi.org/10.1016/s0969-2126(00)00164-7
- Primary Citation of Related Structures:
1EEX, 1EGM, 1EGV - PubMed Abstract:
Adenosylcobalamin (coenzyme B(12)) serves as a cofactor for enzymatic radical reactions. The adenosyl radical, a catalytic radical in these reactions, is formed by homolysis of the cobalt-carbon bond of the coenzyme, although the mechanism of cleavage of its organometallic bond remains unsolved. We determined the three-dimensional structures of diol dehydratase complexed with adeninylpentylcobalamin and with cyanocobalamin at 1.7 A and 1.9 A resolution, respectively, at cryogenic temperatures. In the adeninylpentylcobalamin complex, the adenine ring is bound parallel to the corrin ring as in the free form and methylmalonyl-CoA-mutase-bound coenzyme, but with the other side facing pyrrole ring C. All of its nitrogen atoms except for N(9) are hydrogen-bonded to mainchain amide oxygen and amide nitrogen atoms, a sidechain hydroxyl group, and a water molecule. As compared with the cyanocobalamin complex, the sidechain of Seralpha224 rotates by 120 degrees to hydrogen bond with N(3) of the adenine ring. The structure of the adenine-ring-binding site provides a molecular basis for the strict specificity of diol dehydratase for the coenzyme adenosyl group. The superimposition of the structure of the free coenzyme on that of enzyme-bound adeninylpentylcobalamin demonstrated that the tight enzyme-coenzyme interactions at both the cobalamin moiety and adenine ring of the adenosyl group would inevitably lead to cleavage of the cobalt-carbon bond. Rotation of the ribose moiety around the glycosidic linkage makes the 5'-carbon radical accessible to the hydrogen atom of the substrate to be abstracted.
- Department of Life Science, Himeji Institute of Technology, Kamigori, Akogun, 678-1297, Japan.
Organizational Affiliation: