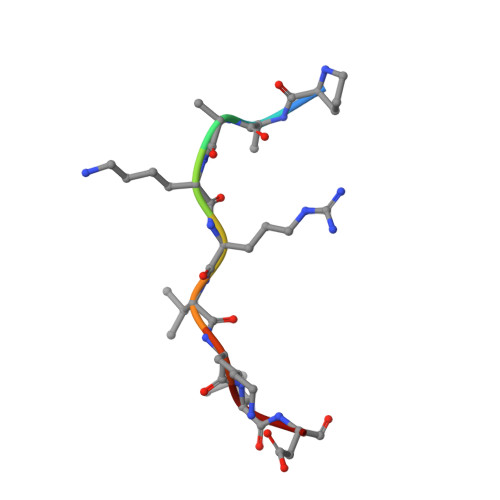
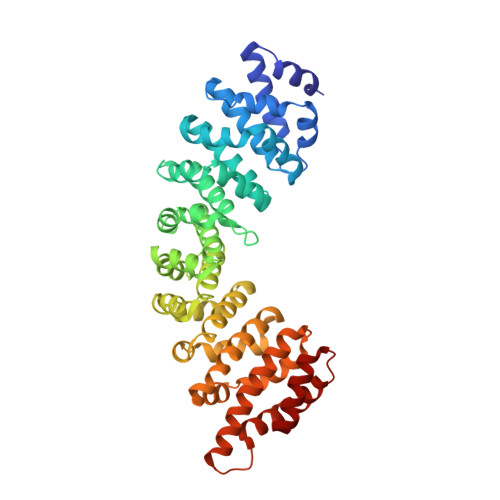Crystallographic analysis of the specific yet versatile recognition of distinct nuclear localization signals by karyopherin alpha.
Conti, E., Kuriyan, J.(2000) Structure 8: 329-338
- PubMed: 10745017
- DOI: https://doi.org/10.1016/s0969-2126(00)00107-6
- Primary Citation of Related Structures:
1EE4, 1EE5 - PubMed Abstract:
Karyopherin alpha (importin alpha) is an adaptor molecule that recognizes proteins containing nuclear localization signals (NLSs). The prototypical NLS that is able to bind to karyopherin alpha is that of the SV40 T antigen, and consists of a short positively charged sequence motif. Distinct classes of NLSs (monopartite and bipartite) have been identified that are only partly conserved with respect to one another but are nevertheless recognized by the same receptor. We report the crystal structures of two peptide complexes of yeast karyopherin alpha (Kapalpha): one with a human c-myc NLS peptide, determined at 2.1 A resolution, and one with a Xenopus nucleoplasmin NLS peptide, determined at 2.4 A resolution. Analysis of these structures reveals the determinants of specificity for the binding of a relatively hydrophobic monopartite NLS and of a bipartite NLS peptide. The peptides bind Kapalpha in its extended surface groove, which presents a modular array of tandem binding pockets for amino acid residues. Monopartite and bipartite NLSs bind to a different number of amino acid binding pockets and make different interactions within them. The relatively hydrophobic monopartite c-myc NLS binds extensively at a few binding pockets in a similar manner to that of the SV40 T antigen NLS. In contrast, the bipartite nucleoplasmin NLS engages the whole array of pockets with individually more limited but overall more abundant interactions, which include the NLS two basic clusters and the backbone of its non-conserved linker region. Versatility in the specific recognition of NLSs relies on the modular.
- Laboratory of Molecular Biophysics, Howard Hughes Medical Institute, The Rockefeller University, New York 10021, USA. conti@embl-heidelberg.de
Organizational Affiliation: