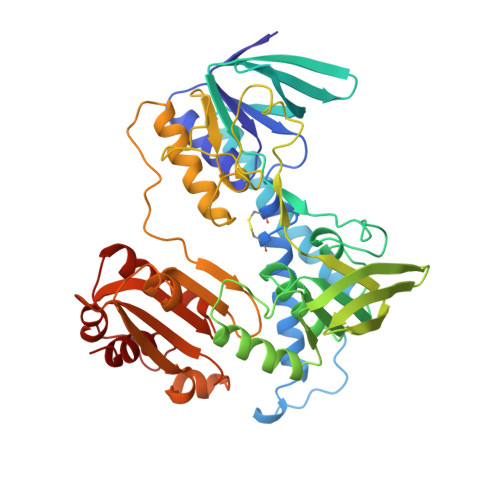
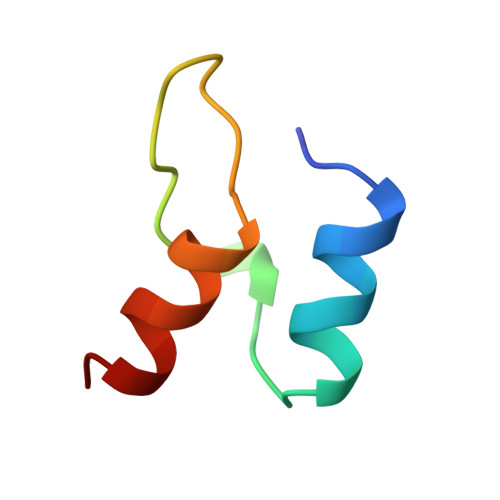
Protein-protein interactions in the pyruvate dehydrogenase multienzyme complex: dihydrolipoamide dehydrogenase complexed with the binding domain of dihydrolipoamide acetyltransferase.
Mande, S.S., Sarfaty, S., Allen, M.D., Perham, R.N., Hol, W.G.(1996) Structure 4: 277-286
- PubMed: 8805537
- DOI: https://doi.org/10.1016/s0969-2126(96)00032-9
- Primary Citation of Related Structures:
1EBD - PubMed Abstract:
The ubiquitous pyruvate dehydrogenase multienzyme complex is built around an octahedral or icosahedral core of dihydrolipoamide acetyltransferase (E2) chains, to which multiple copies of pyruvate decarboxylase (E1) and dihydrolipoamide dehydrogenase (E3) bind tightly but non-covalently. E2 is a flexible multidomain protein that mediates interactions with E1 and E3 through a remarkably small binding domain (E2BD). In the Bacillus stearothermophilus complex, the E2 core is an icosahedral assembly of 60 E2 chains. The crystal structure of the E3 dimer (101 kDa) complexed with E2BD (4 kDa) has been solved to 2.6 A resolution. Interactions between E3 and E2BD are dominated by an electrostatic zipper formed by Arg135 and Arg139 in the N-terminal helix of E2BD and Asp344 and Glu431 of one of the monomers of E3. E2BD interacts with both E3 monomers, but the binding site is located close to the twofold axis. Thus, in agreement with earlier biochemical results, it is impossible for two molecules of E2BD to bind simultaneously to one E3 dimer. Combining this new structure for the E3-E2BD complex with previously determined structures of the E2 catalytic domain and the E2 lipoyl domain creates a model of the E2 core showing how the lipoyl domain can move between the active sites of E2 and E3 in the multienzyme complex.
- Department of Biological Structure, Biomolecular Structure Center, University of Washington, Box 357742, Seattle, WA 98195, USA.
Organizational Affiliation: