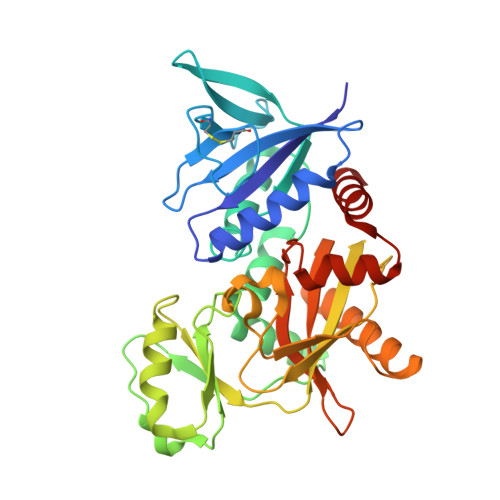
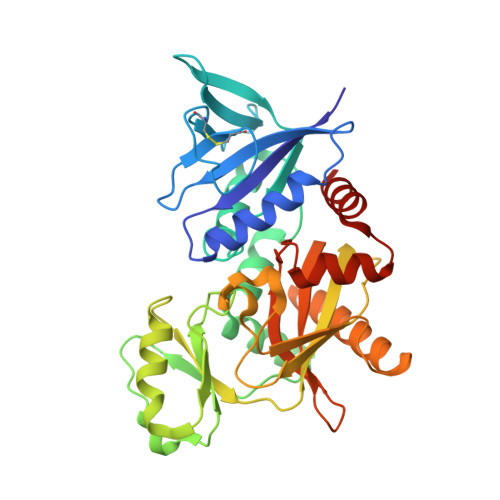
The molecular basis of vancomycin resistance in clinically relevant Enterococci: crystal structure of D-alanyl-D-lactate ligase (VanA).
Roper, D.I., Huyton, T., Vagin, A., Dodson, G.(2000) Proc Natl Acad Sci U S A 97: 8921-8925
- PubMed: 10908650
- DOI: https://doi.org/10.1073/pnas.150116497
- Primary Citation of Related Structures:
1E4E - PubMed Abstract:
d-alanine-d-lactate ligase from Enterococcus faecium BM4147 is directly responsible for the biosynthesis of alternate cell-wall precursors in bacteria, which are resistant to the glycopeptide antibiotic vancomycin. The crystal structure has been determined with data extending to 2.5-A resolution. This structure shows that the active site has unexpected interactions and is distinct from previous models for d-alanyl-d-lactate ligase mechanistic studies. It appears that the preference of the enzyme for lactate as a ligand over d-alanine could be mediated by electrostatic effects and/or a hydrogen-bonding network, which principally involve His-244. The structure of d-alanyl-d-lactate ligase provides a revised interpretation of the molecular events that lead to vancomycin resistance.
- York Structural Biology Laboratory, Department of Chemistry, The University of York, Heslington, York YO10 5DD, United Kingdom. roper@ysbl.york.ac.uk
Organizational Affiliation: