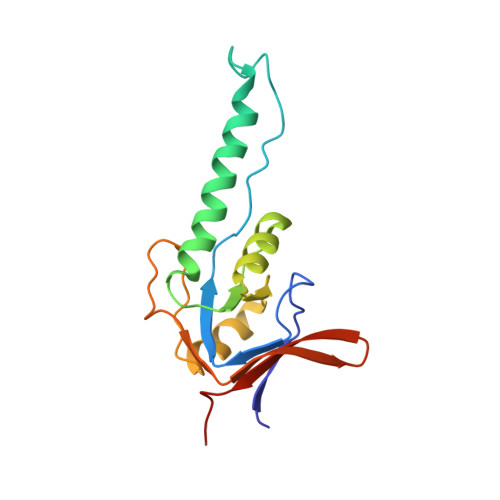
Crystal Structure of the Beta-Apical Domain from Thermosome Reveals Structural Plasticity in Protrusion Region
Bosch, G., Baumeister, W., Essen, L.-O.(2000) J Mol Biology 301: 19
- PubMed: 10926489
- DOI: https://doi.org/10.1006/jmbi.2000.3955
- Primary Citation of Related Structures:
1E0R - PubMed Abstract:
The crystal structure of the beta-apical domain of the thermosome, an archaeal group II chaperonin from Thermoplasma acidophilum, has been determined at 2.8 A resolution. The structure shows an invariant globular core from which a 25 A long protrusion emanates, composed of an elongated alpha-helix (H10) and a long extended stretch consisting of residues GluB245-ThrB253. A comparison with previous apical domain structures reveals a large segmental displacement of the protruding part of helix H10 via the hinge GluB276-ValB278. The region comprising residues GluB245-ThrB253 adopts an extended beta-like conformation rather than the alpha-helix seen in the alpha-apical domain. Consequently, it appears that the protrusions of the apical domains from group II chaperonins might assume a variety of context-dependent conformations during an open, substrate-accepting state of the chaperonin. Sequence variations in the protrusion regions that are found in the eukaryotic TRiC/CCT subunits may provide different structural propensities and hence serve different roles in substrate recognition.
- Max-Planck-Institute for Biochemistry, Am Klopferspitz 18a, Martinsried bei München, D-82152, Germany.
Organizational Affiliation: