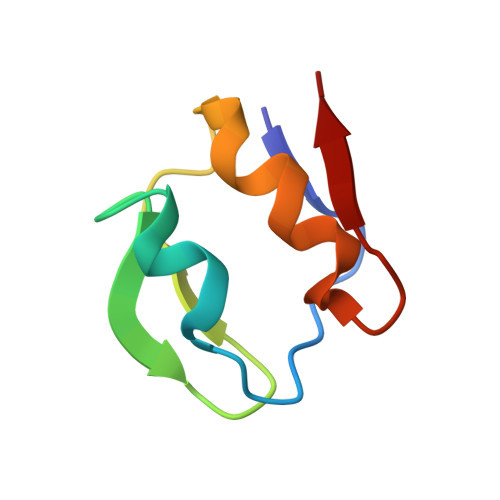
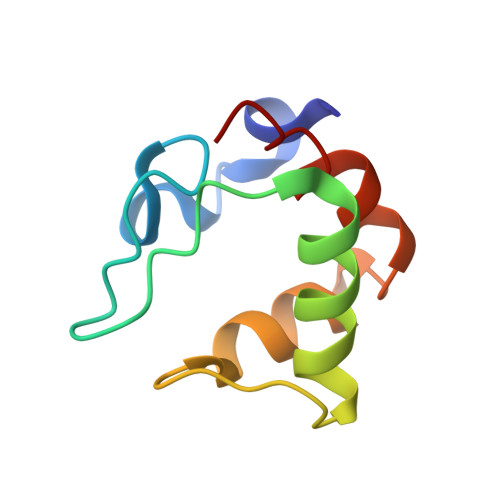
Heteronuclear NMR and Soft Docking: An Experimental Approach for a Structural Model of the Cytochrome C553-Ferredoxin Complex
Morelli, X., Dolla, A., Czjzek, M., Palma, P.N., Blasco, F., Krippahl, L., Moura, J.J.G., Guerlesquin, F.(2000) Biochemistry 39: 2530
- PubMed: 10704202
- DOI: https://doi.org/10.1021/bi992306s
- Primary Citation of Related Structures:
1DWL - PubMed Abstract:
The combination of docking algorithms with NMR data has been developed extensively for the studies of protein-ligand interactions. However, to extend this development for the studies of protein-protein interactions, the intermolecular NOE constraints, which are needed, are more difficult to access. In the present work, we describe a new approach that combines an ab initio docking calculation and the mapping of an interaction site using chemical shift variation analysis. The cytochrome c553-ferredoxin complex is used as a model of numerous electron-transfer complexes. The 15N-labeling of both molecules has been obtained, and the mapping of the interacting site on each partner, respectively, has been done using HSQC experiments. 1H and 15N chemical shift analysis defines the area of both molecules involved in the recognition interface. Models of the complex were generated by an ab initio docking software, the BiGGER program (bimolecular complex generation with global evaluation and ranking). This program generates a population of protein-protein docked geometries ranked by a scoring function, combining relevant stabilization parameters such as geometric complementarity surfaces, electrostatic interactions, desolvation energy, and pairwise affinities of amino acid side chains. We have implemented a new module that includes experimental input (here, NMR mapping of the interacting site) as a filter to select the accurate models. Final structures were energy minimized using the X-PLOR software and then analyzed. The best solution has an interface area (1037.4 A2) falling close to the range of generally observed recognition interfaces, with a distance of 10.0 A between the redox centers.
- Unité de Bioénergétique et Ingénierie des Protéines, IBSM-CNRS, 31 chemin Joseph Aiguier, 13402 Marseille cedex 20, France.
Organizational Affiliation: