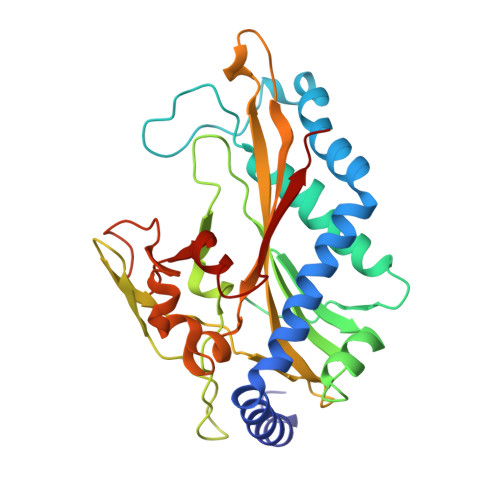
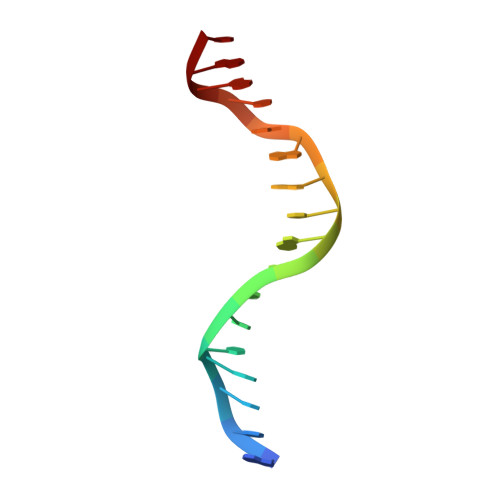
Crystal structure of restriction endonuclease BglI bound to its interrupted DNA recognition sequence.
Newman, M., Lunnen, K., Wilson, G., Greci, J., Schildkraut, I., Phillips, S.E.(1998) EMBO J 17: 5466-5476
- PubMed: 9736624
- DOI: https://doi.org/10.1093/emboj/17.18.5466
- Primary Citation of Related Structures:
1DMU - PubMed Abstract:
The crystal structure of the type II restriction endonuclease BglI bound to DNA containing its specific recognition sequence has been determined at 2.2 A resolution. This is the first structure of a restriction endonuclease that recognizes and cleaves an interrupted DNA sequence, producing 3' overhanging ends. BglI is a homodimer that binds its specific DNA sequence with the minor groove facing the protein. Parts of the enzyme reach into both the major and minor grooves to contact the edges of the bases within the recognition half-sites. The arrangement of active site residues is strikingly similar to other restriction endonucleases, but the co-ordination of two calcium ions at the active site gives new insight into the catalytic mechanism. Surprisingly, the core of a BglI subunit displays a striking similarity to subunits of EcoRV and PvuII, but the dimer structure is dramatically different. The BglI-DNA complex demonstrates, for the first time, that a conserved subunit fold can dimerize in more than one way, resulting in different DNA cleavage patterns.
- School of Biochemistry and Molecular Biology, and North of England Structural Biology Centre, University of Leeds, Leeds LS2 9JT, UK.
Organizational Affiliation: