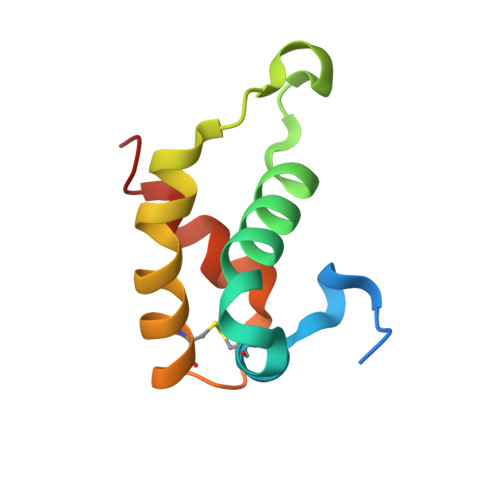
HDEA, a periplasmic protein that supports acid resistance in pathogenic enteric bacteria.
Gajiwala, K.S., Burley, S.K.(2000) J Mol Biology 295: 605-612
- PubMed: 10623550
- DOI: https://doi.org/10.1006/jmbi.1999.3347
- Primary Citation of Related Structures:
1DJ8 - PubMed Abstract:
The X-ray crystal structure of the Escherichia coli stress response protein HDEA has been determined at 2.0 A resolution. The single domain alpha-helical protein is found in the periplasmic space, where it supports an acid resistance phenotype essential for infectivity of enteric bacterial pathogens, such as Shigella and E. coli. Functional studies demonstrate that HDEA is activated by a dimer-to-monomer transition at acidic pH, leading to suppression of aggregation by acid-denatured proteins. We suggest that HDEA may support chaperone-like functions during the extremely acidic conditions.
- Laboratorie of Molecular Biophysics, Howard Hughes Medical Institute, New York, 10021, USA.
Organizational Affiliation: