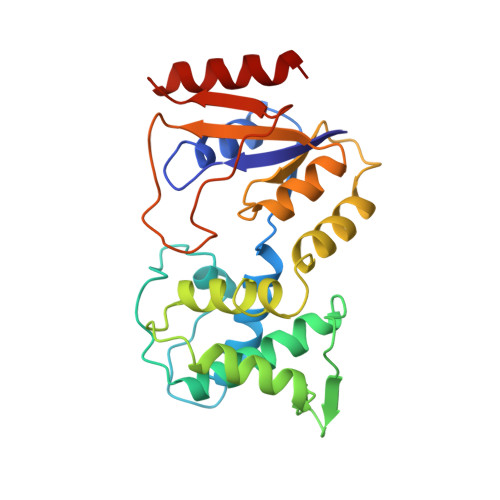
Crystal structure of bacteriophage T4 deoxynucleotide kinase with its substrates dGMP and ATP.
Teplyakov, A., Sebastiao, P., Obmolova, G., Perrakis, A., Brush, G.S., Bessman, M.J., Wilson, K.S.(1996) EMBO J 15: 3487-3497
- PubMed: 8670851
- Primary Citation of Related Structures:
1DEK, 1DEL - PubMed Abstract:
NMP kinases catalyse the phosphorylation of the canonical nucleotides to the corresponding diphosphates using ATP as a phosphate donor. Bacteriophage T4 deoxynucleotide kinase (DNK) is the only member of this family of enzymes that recognizes three structurally dissimilar nucleotides: dGMP, dTMP and 5-hydroxymethyl-dCMP while excluding dCMP and dAMP. The crystal structure of DNK with its substrate dGMP has been determined at 2.0 A resolution by single isomorphous replacement. The structure of the ternary complex with dGMP and ATP has been determined at 2.2 A resolution. The polypeptide chain of DNK is folded into two domains of equal size, one of which resembles the mononucleotide binding motif with the glycine-rich P-loop. The second domain, consisting of five alpha-helices, forms the NMP binding pocket. A hinge connection between the domains allows for large movements upon substrate binding which are not restricted by dimerization of the enzyme. The mechanism of active centre formation via domain closure is described. Comparison with other P-loop-containing proteins indicates an induced-fit mode of NTP binding. Protein-substrate interactions observed at the NMP and NTP sites provide the basis for understanding the principles of nucleotide discrimination.
- European Molecular Biology Laboratory, DESY, Hamburg, Germany.
Organizational Affiliation: