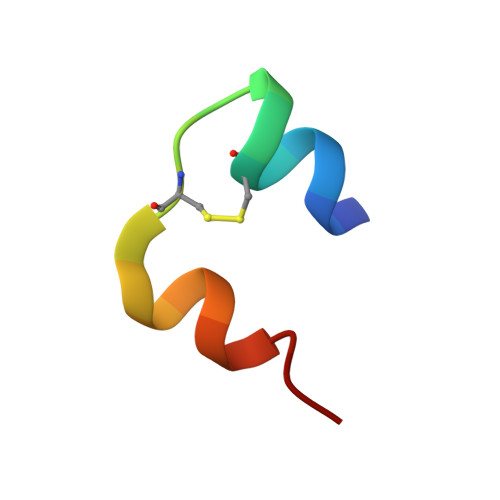
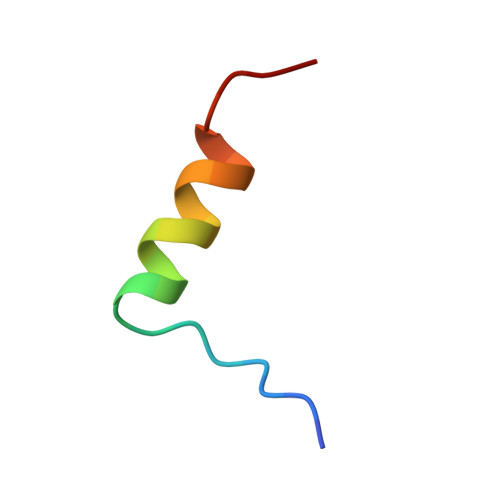
Crystal structure of desheptapeptide(B24-B30)insulin at 1.6 A resolution: implications for receptor binding.
Bao, S.J., Xie, D.L., Zhang, J.P., Chang, W.R., Liang, D.C.(1997) Proc Natl Acad Sci U S A 94: 2975-2980
- PubMed: 9096331
- DOI: https://doi.org/10.1073/pnas.94.7.2975
- Primary Citation of Related Structures:
1DEI - PubMed Abstract:
The crystal structure of desheptapeptide (B24-B30) insulin (DHPI), a virtually inactive analog of insulin, was determined at 1.6 A resolution. In the refined structure model, DHPI retains three alpha-helices (A1-A8, A12-A18, and B9-B19) as its structural framework, while great conformational changes occur in the N and C termini of B-chain. The beta-turn, which lies in B20-B30 in insulin and insulin analogs with high potency, no longer exists in DHPI. Relative motion is observed among the three alpha-helices, each as a rigid functional group. In contrast, a region covering B5-B6 and A6-A11 exhibits a relatively stable conformation. We interpret our results as identifying: (i) the importance of beta-turn in determining the receptor-binding potency of insulin and (ii) a leading role of PheB24 in maintaining the beta-turn structure.
- National Laboratory of Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing.
Organizational Affiliation: