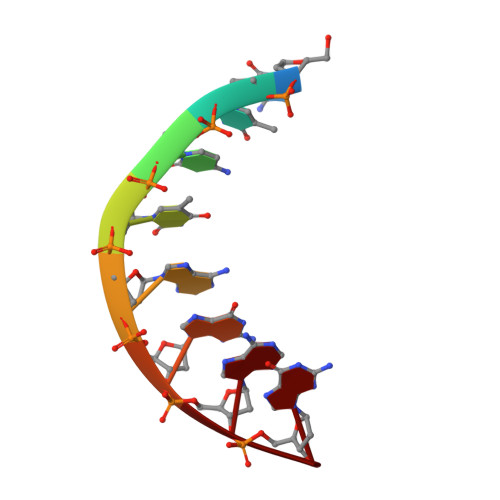
Structural variation in d(CTCTAGAG). Implications for protein-DNA interactions.
Hunter, W.N., D'Estaintot, B.L., Kennard, O.(1989) Biochemistry 28: 2444-2451
- PubMed: 2730875
- DOI: https://doi.org/10.1021/bi00432a015
- Primary Citation of Related Structures:
1D93 - PubMed Abstract:
Single-crystal X-ray diffraction techniques have been used to characterize the structure of the self-complementary DNA oligomer d(CTCTAGAG). The structure was refined to an R factor of 14.7% using data to 2.15-A resolution. The tetragonal unit cell, space group P4(3)2(1)2, has dimensions a = 42.53 and c = 24.33 A. The asymmetric unit consists of a single strand or four base pairs. Two strands, related by a crystallographic dyad axis, coil about each other to form a right-handed duplex. This octamer duplex has a mean helix rotation of 32 degrees, 11.3 base pairs per turn, an average rise of 3.1 A, C3'-endo furanose conformations, a shallow minor groove, and a deep major groove. Such averaged parameters suggest classification of the octamer as a member of the A-DNA family. However, the global parameters tend to mask variations in conformational parameters observed at the level of the base pairs. In particular, the central TpA (= TpA) step displays extensive interstrand purine-purine overlap and an unusual sugar-phosphate backbone conformation. These structural features may be directly related to certain sequence-specific protein-DNA interactions involving nucleases and repressors.
- University Chemical Laboratory, Cambridge, U.K.
Organizational Affiliation: