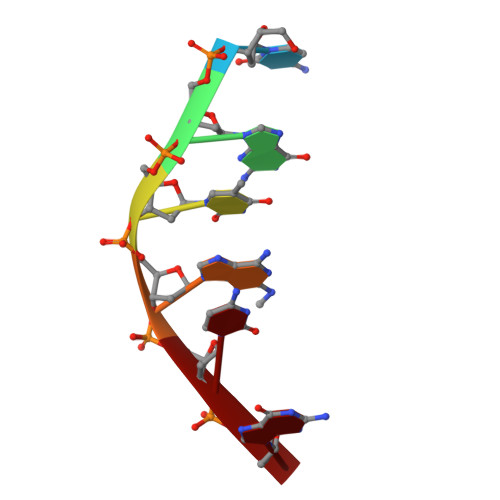
Facile formation of a crosslinked adduct between DNA and the daunorubicin derivative MAR70 mediated by formaldehyde: molecular structure of the MAR70-d(CGTnACG) covalent adduct.
Gao, Y.G., Liaw, Y.C., Li, Y.K., van der Marel, G.A., van Boom, J.H., Wang, A.H.(1991) Proc Natl Acad Sci U S A 88: 4845-4849
- PubMed: 2052564
- DOI: https://doi.org/10.1073/pnas.88.11.4845
- Primary Citation of Related Structures:
1D35, 1D36 - PubMed Abstract:
MAR70 is a synthetic derivative of the anticancer drug daunorubicin that contains an additional sugar, attached to the O4' of daunosamine. When MAR70 was crystallized with the DNA hexamer d(CGTnACG), where nA is 2-aminoadenine, a covalent methylene bridge was formed between the N3' of daunosamine and the N2 of 2-aminoadenine. This spontaneous reaction occurred through the crosslinking action of formaldehyde. The crosslink was demonstrated by the three-dimensional structure of the 2:1 adduct between MAR70 and d(CGTnACG) solved at 1.3-A resolution by x-ray diffraction analysis. The perfect juxtaposition of the two amino groups in the complex provides a template for efficient addition of formaldehyde. This adduct structure is compared with the analogous structure at 1.5-A resolution of the complex of MAR70-d(CGTACG), in which no formaldehyde addition was observed. In both complexes, two MAR70 molecules bind to the DNA hexamer double helix; the elongated aglycon chromophore is intercalated between the CpG steps and spans the G.C Watson-Crick base pairs. The disaccharides occupy nearly the entire minor groove of the distorted B-DNA hexamer double helix. The second sugar is in contact with the sugar-phosphate backbone and does not affect the binding interactions of the daunorubicin portion to DNA. The structure allows us to model the binding to DNA of drugs having more extensive oligosaccharides. In addition, it suggests that placing a reactive (e.g., alkylating) functional group at the N3' amino position of daunorubicin might be a fruitful route for designing anticancer drugs.
- Department of Physiology and Biophysics, University of Illinois, Urbana-Champaign 61801.
Organizational Affiliation: