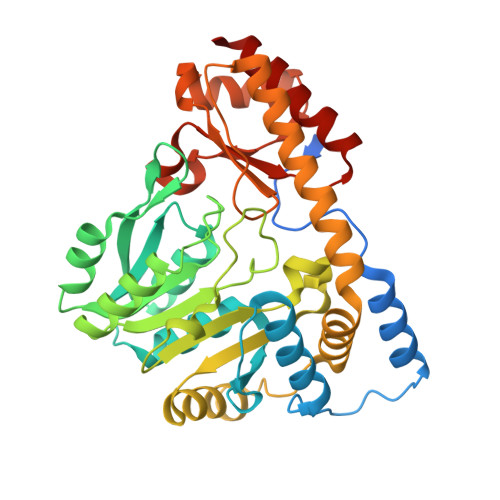
X-ray structure of MalY from Escherichia coli: a pyridoxal 5'-phosphate-dependent enzyme acting as a modulator in mal gene expression.
Clausen, T., Schlegel, A., Peist, R., Schneider, E., Steegborn, C., Chang, Y.S., Haase, A., Bourenkov, G.P., Bartunik, H.D., Boos, W.(2000) EMBO J 19: 831-842
- PubMed: 10698925
- DOI: https://doi.org/10.1093/emboj/19.5.831
- Primary Citation of Related Structures:
1D2F - PubMed Abstract:
MalY represents a bifunctional pyridoxal 5'-phosphate-dependent enzyme acting as a beta-cystathionase and as a repressor of the maltose regulon. Here we present the crystal structures of wild-type and A221V mutant protein. Each subunit of the MalY dimer is composed of a large pyridoxal 5'-phosphate-binding domain and a small domain similar to aminotransferases. The structural alignment with related enzymes identifies residues that are generally responsible for beta-lyase activity and depicts a unique binding mode of the pyridoxal 5'-phosphate correlated with a larger, more flexible substrate-binding pocket. In a screen for MalY mutants with reduced mal repressor properties, mutations occurred in three clusters: I, 83-84; II, 181-189 and III, 215-221, which constitute a clearly distinguished region in the MalY crystal structure far away from the cofactor. The tertiary structure of one of these mutants (A221V) demonstrates that positional rearrangements are indeed restricted to regions I, II and III. Therefore, we propose that a direct protein-protein interaction with MalT, the central transcriptional activator of the maltose system, underlies MalY-dependent repression of the maltose system.
- Max-Planck-Institut für Biochemie, Abteilung Strukturforschung, D-82152 Martinsried.
Organizational Affiliation: