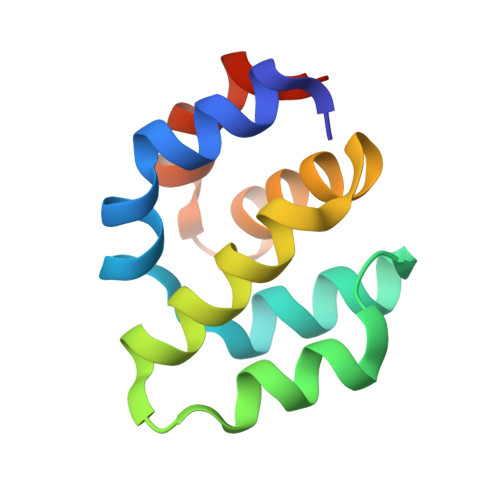
Crystal structure of Apaf-1 caspase recruitment domain: an alpha-helical Greek key fold for apoptotic signaling.
Vaughn, D.E., Rodriguez, J., Lazebnik, Y., Joshua-Tor, L.(1999) J Mol Biology 293: 439-447
- PubMed: 10543941
- DOI: https://doi.org/10.1006/jmbi.1999.3177
- Primary Citation of Related Structures:
1CY5 - PubMed Abstract:
The caspase recruitment domain (CARD) of Apaf-1 binds to the CARD of caspase-9 to trigger a proteolytic cascade that leads to apoptotic cell death. We report the crystal structure of the Apaf-1 CARD at 1. 3 A resolution, solved in a two-element multiwavelength anomalous dispersion (MAD) X-ray diffraction experiment. This CARD adopts a six-helix bundle fold with Greek key topology surrounding an extensive hydrophobic core. This fold, which we call the "death fold", is found in other domains that mediate interactions in apoptotic signaling despite very low sequence identity. From a structure-based alignment, we identify conserved patterns that characterize the death fold and its subclasses. Like the Ig-fold, it provides a rigid structural scaffold upon which diverse recognition surfaces are assembled.
- W. M. Keck Structural Biology, Cold Spring Harbor, NY 11724, USA.
Organizational Affiliation: