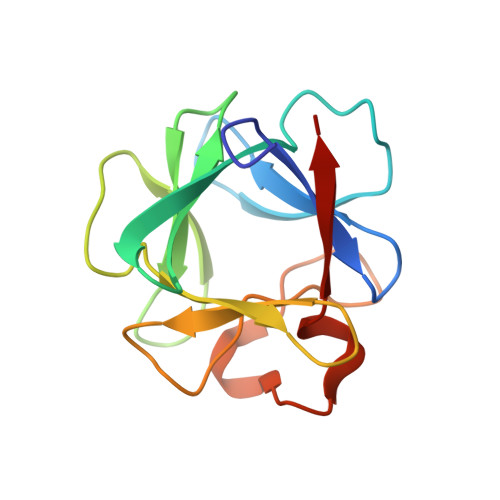
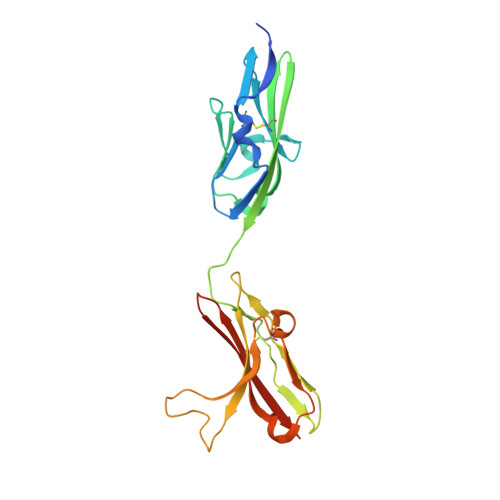
Structural basis for FGF receptor dimerization and activation.
Plotnikov, A.N., Schlessinger, J., Hubbard, S.R., Mohammadi, M.(1999) Cell 98: 641-650
- PubMed: 10490103
- DOI: https://doi.org/10.1016/s0092-8674(00)80051-3
- Primary Citation of Related Structures:
1CVS - PubMed Abstract:
The crystal structure of FGF2 bound to a naturally occurring variant of FGF receptor 1 (FGFR1) consisting of immunoglobulin-like domains 2 (D2) and 3 (D3) has been determined at 2.8 A resolution. Two FGF2:FGFR1 complexes form a 2-fold symmetric dimer. Within each complex, FGF2 interacts extensively with D2 and D3 as well as with the linker between the two domains. The dimer is stabilized by interactions between FGF2 and D2 of the adjoining complex and by a direct interaction between D2 of each receptor. A positively charged canyon formed by a cluster of exposed basic residues likely represents the heparin-binding site. A general model for FGF- and heparin-induced FGFR dimerization is inferred from the crystal structure, unifying a wealth of biochemical data.
- Department of Pharmacology, New York University School of Medicine, New York 10016, USA.
Organizational Affiliation: