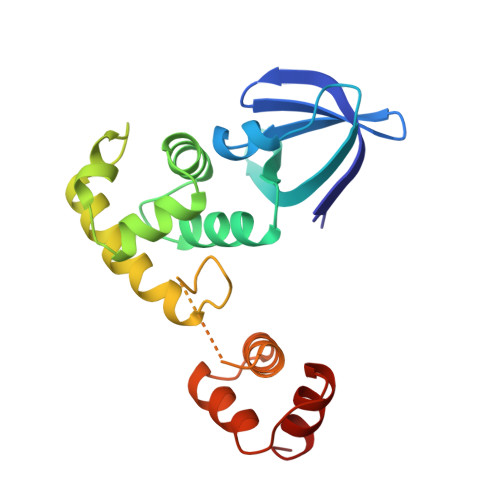
Crystal structure of DNA recombination protein RuvA and a model for its binding to the Holliday junction.
Rafferty, J.B., Sedelnikova, S.E., Hargreaves, D., Artymiuk, P.J., Baker, P.J., Sharples, G.J., Mahdi, A.A., Lloyd, R.G., Rice, D.W.(1996) Science 274: 415-421
- PubMed: 8832889
- DOI: https://doi.org/10.1126/science.274.5286.415
- Primary Citation of Related Structures:
1CUK - PubMed Abstract:
The Escherichia coli DNA binding protein RuvA acts in concert with the helicase RuvB to drive branch migration of Holliday intermediates during recombination and DNA repair. The atomic structure of RuvA was determined at a resolution of 1.9 angstroms. Four monomers of RuvA are related by fourfold symmetry in a manner reminiscent of a four-petaled flower. The four DNA duplex arms of a Holliday junction can be modeled in a square planar configuration and docked into grooves on the concave surface of the protein around a central pin that may facilitate strand separation during the migration reaction. The model presented reveals how a RuvAB-junction complex may also accommodate the resolvase RuvC.
- Krebs Institute, Department of Molecular Biology and Biotechnology, University of Sheffield, Western Bank, Sheffield S10 2TN, UK. d.rice@sheffield.ac.uk
Organizational Affiliation: