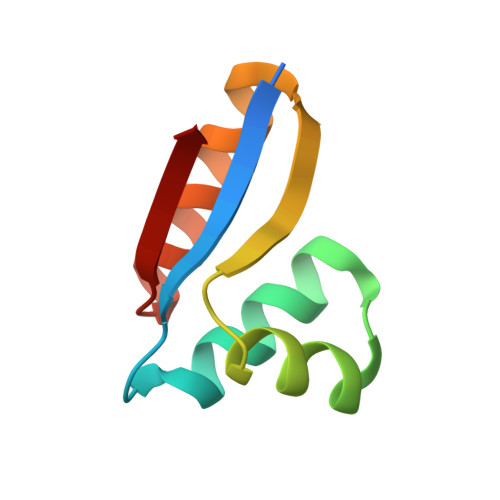
Structure of the C-terminal domain of the ribosomal protein L7/L12 from Escherichia coli at 1.7 A.
Leijonmarck, M., Liljas, A.(1987) J Mol Biology 195: 555-579
- PubMed: 3309338
- DOI: https://doi.org/10.1016/0022-2836(87)90183-5
- Primary Citation of Related Structures:
1CTF - PubMed Abstract:
The structure of a C-terminal fragment of the ribosomal protein L7/L12 from Escherichia coli has been refined using crystallographic data to 1.7 A resolution. The R-value is 17.4%. Six residues at the N terminus are too disordered in the structure to be localized. These residues are probably part of a hinge in the complete L7/L12 molecule. The possibility that a 2-fold crystallographic axis is a molecular 2-fold axis is discussed. A patch of invariant residues on the surface of the dimer is probably involved in functional interactions with elongation factors.
- Institute of Molecular Biology, University of Uppsala, Sweden.
Organizational Affiliation: