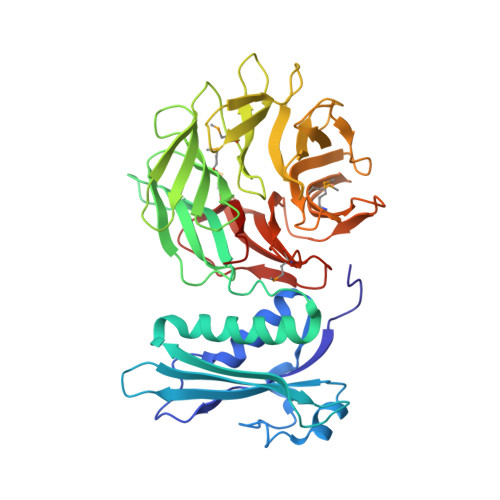
Structure of the Escherichia coli TolB protein determined by MAD methods at 1.95 A resolution.
Abergel, C., Bouveret, E., Claverie, J.M., Brown, K., Rigal, A., Lazdunski, C., Benedetti, H.(1999) Structure 7: 1291-1300
- PubMed: 10545334
- DOI: https://doi.org/10.1016/s0969-2126(00)80062-3
- Primary Citation of Related Structures:
1CRZ - PubMed Abstract:
The periplasmic protein TolB from Escherichia coli is part of the Tol-PAL (peptidoglycan-associated lipoprotein) multiprotein complex used by group A colicins to penetrate and kill cells. TolB homologues are found in many gram-negative bacteria and the Tol-PAL system is thought to play a role in bacterial envelope integrity. TolB is required for lethal infection by Salmonella typhimurium in mice. The crystal structure of the selenomethionine-substituted TolB protein from E. coli was solved using multiwavelength anomalous dispersion methods and refined to 1. 95 A. TolB has a two-domain structure. The N-terminal domain consists of two alpha helices, a five-stranded beta-sheet floor and a long loop at the back of this floor. The C-terminal domain is a six-bladed beta propeller. The small, possibly mobile, contact area (430 A(2)) between the two domains involves residues from the two helices and the first and sixth blades of the beta propeller. All available genomic sequences were used to identify new TolB homologues in gram-negative bacteria. The TolB structure was then interpreted using the observed conservation pattern. The TolB beta-propeller C-terminal domain exhibits sequence similarities to numerous members of the prolyl oligopeptidase family and, to a lesser extent, to class B metallo-beta-lactamases. The alpha/beta N-terminal domain shares a structural similarity with the C-terminal domain of transfer RNA ligases. We suggest that the TolB protein might be part of a multiprotein complex involved in the recycling of peptidoglycan or in its covalent linking with lipoproteins.
- Information Génétique et Structurale, CNRS-UMR 1889 Institut de Biologie Structurale et Microbiologie 31 Chemin Joseph Aiguier, Marseille, 13402, Cedex 20, France. chantal@igs.cnrs-mrs.fr
Organizational Affiliation: