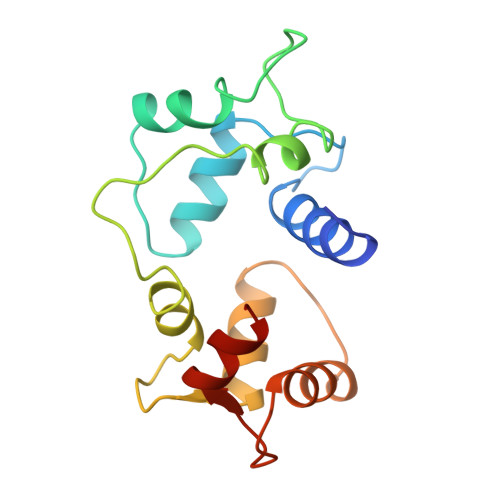
Motions of calmodulin characterized using both Bragg and diffuse X-ray scattering.
Wall, M.E., Clarage, J.B., Phillips Jr., G.N.(1997) Structure 5: 1599-1612
- PubMed: 9438860
- DOI: https://doi.org/10.1016/s0969-2126(97)00308-0
- Primary Citation of Related Structures:
1CM1, 1CM4 - PubMed Abstract:
Calmodulin is a calcium-activated regulatory protein which can bind to many different targets. The protein resembles a highly flexible dumbbell, and bends in the middle as it binds. This and other motions must be understood to formulate a realistic model of calmodulin function. Using the Bragg reflections from X-ray crystallography, a multiple-conformer refinement of a calmodulin-peptide complex shows anisotropic displacements, with high variations of dihedral angles in several nonhelical domains: the flexible linker; three of the four calcium-binding sites (including both of the N-terminal sites); and a turn connecting the C-terminal EF-hand calcium-binding domains. Three-dimensional maps of the large scale diffuse X-ray scattering data show isotropic liquid-like motions with an unusually small correlation length. Three-dimensional maps of the small scale diffuse streaks show highly coupled, anisotropic motions along the head-to-tail molecular packing direction in the unit cell. There is also weak coupling perpendicular to the head-to-tail packing direction, particularly across a cavity occupied by the disordered linker domain of the molecule. Together, the Bragg and diffuse scattering present a self-consistent description of the motions in the flexible linker of calmodulin. The other mobile regions of the protein are also of great interest. In particular, the high variations in the calcium-binding sites are likely to influence how strongly they bind ions. This is especially important in the N-terminal sites, which regulate the activity of the molecule.
- Department of Biochemistry and Cell Biology, The WM Keck Center for Computational Biology, Rice University Houston, TX 77005-1892, USA, mewall@bioc.rice.edu
Organizational Affiliation: