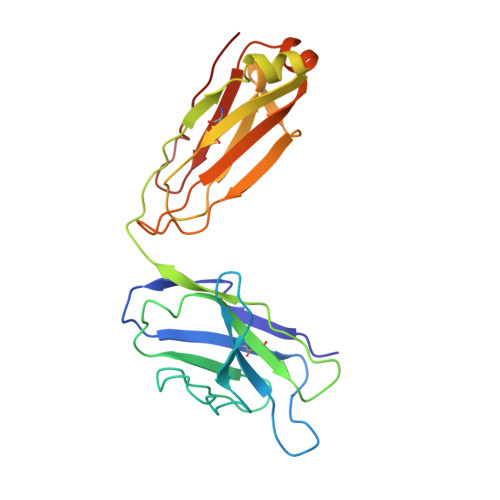
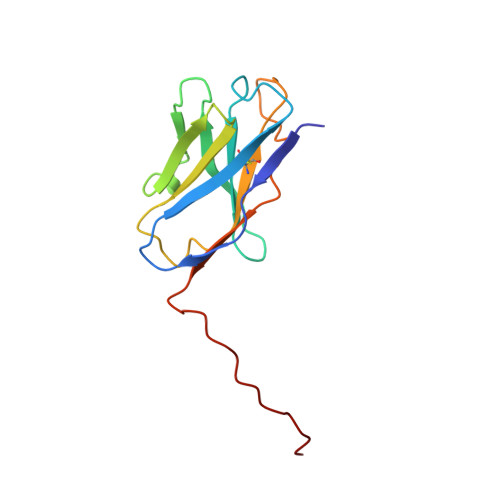
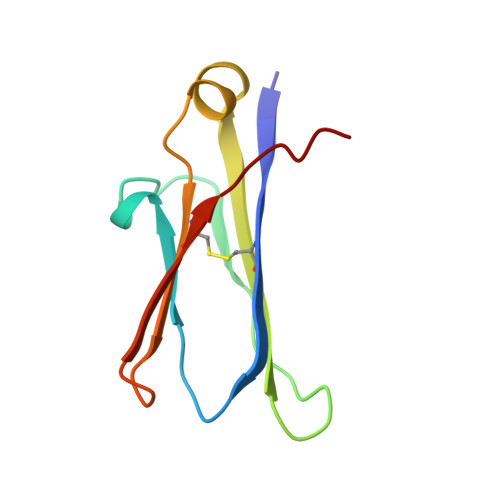
Inhibition of the HIV-1 and HIV-2 proteases by a monoclonal antibody.
Lescar, J., Brynda, J., Rezacova, P., Stouracova, R., Riottot, M.M., Chitarra, V., Fabry, M., Horejsi, M., Sedlacek, J., Bentley, G.A.(1999) Protein Sci 8: 2686-2696
- PubMed: 10631984
- DOI: https://doi.org/10.1110/ps.8.12.2686
- Primary Citation of Related Structures:
1CL7 - PubMed Abstract:
The monoclonal antibody 1696, directed against the HIV-1 protease, displays strong inhibitory effects toward the catalytic activity of the enzyme of both the HIV-1 and HIV-2 isolates. This antibody cross-reacts with peptides that include the N-terminus of the enzyme, a region that is well conserved in sequence among different viral strains and which, furthermore, is crucial for homodimerization to the active enzymatic form. This observation, as well as antigen-binding studies in the presence of an active site inhibitor, suggest that 1696 inhibits the HIV protease by destabilizing its active homodimeric form. To characterize further how the antibody 1696 inhibits the HIV-1 and HIV-2 proteases, we have solved the crystal structure of its Fab fragment by molecular replacement and refined it at 3.0 A resolution. The antigen binding site has a deep cavity at its center, which is lined mainly by acidic and hydrophobic residues, and is large enough to accommodate several antigen residues. The structure of the Fab 1696 could form a starting basis for the design of alternative HIV protease-inhibiting molecules of broad specificity.
- Unité d'Immunologie Structurale (URA 1961 CNRS), Département d'Immunologie, Institut Pasteur, Paris, France.
Organizational Affiliation: