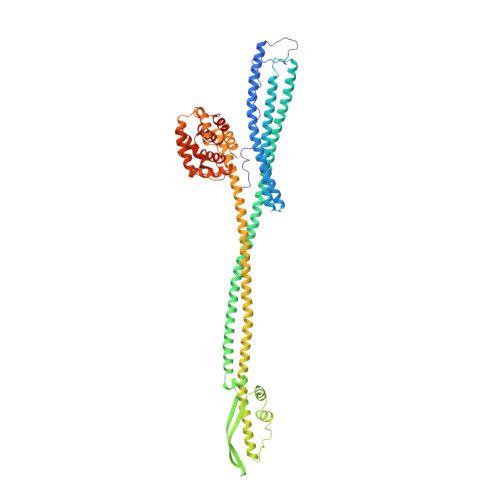
Crystal structure of colicin Ia.
Wiener, M., Freymann, D., Ghosh, P., Stroud, R.M.(1997) Nature 385: 461-464
- PubMed: 9009197
- DOI: https://doi.org/10.1038/385461a0
- Primary Citation of Related Structures:
1CII - PubMed Abstract:
The ion-channel forming colicins A, B, E1, Ia, Ib and N all kill bacterial cells selectively by co-opting bacterial active-transport pathways and forming voltage-gated ion conducting channels across the plasma membrane of the target bacterium. The crystal structure of colicin Ia reveals a molecule 210 A long with three distinct functional domains arranged along a backbone of two extraordinarily long alpha-helices. A central domain at the bend of the hairpin-like structure mediates specific recognition and binding to an outer-membrane receptor. A second domain mediates translocation across the outer membrane via the TonB transport pathway; the TonB-box recognition element of colicin Ia is on one side of three 80 A-long helices arranged as a helical sheet. A third domain is made up of 10 alpha-helices which form a voltage-activated and voltage-gated ion conducting channel across the plasma membrane of the target cell. The two 160 A-long alpha-helices that link the receptor-binding domain to the other domains enable the colicin Ia molecule to span the periplasmic space and contact both the outer and plasma membranes simultaneously during function.
- S-964 Department of Biochemistry and Biophysics, University of California, San Francisco 94143-0448, USA.
Organizational Affiliation: