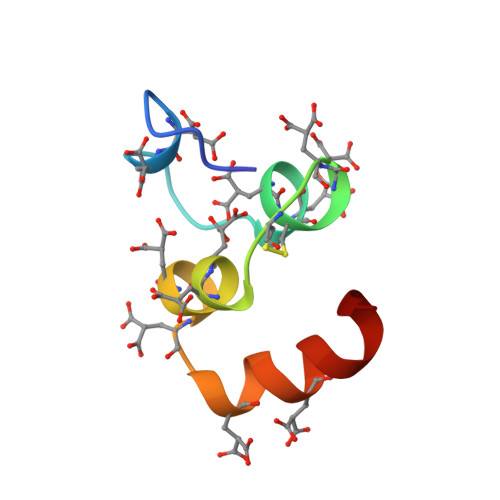
Structure of the calcium ion-bound gamma-carboxyglutamic acid-rich domain of factor IX.
Freedman, S.J., Furie, B.C., Furie, B., Baleja, J.D.(1995) Biochemistry 34: 12126-12137
- PubMed: 7547952
- DOI: https://doi.org/10.1021/bi00038a005
- Primary Citation of Related Structures:
1CFI - PubMed Abstract:
We have determined the Ca(II)-bound structure of factor IX, residues 1-47, by nuclear magnetic resonance (NMR) spectroscopy. The amino-terminal 47 residues include the gamma-carboxyglutamic acid-rich and aromatic amino acid stack domains, and this region is responsible for Ca(II)-dependent phospholipid binding in factor IX. Protons in the 1-47 amino acid sequence were assigned using standard two-dimensional homonuclear NMR experiments. A total of 851 distance restraints and 57 torsion angle restraints were used to generate 17 final structures by distance geometry and simulated annealing methods. The backbone RMSD to the geometric average is 0.6 +/- 0.1 A. The Ca(II)-bound structure is substantially more ordered with increased helical content compared to the apo-factor IX (1-47) structure. The global fold is similar to the crystal structure of the Ca(II)-bound Gla domain of prothrombin fragment I from residues 12 to 47 (RMSD approximately 1.3 A), but the backbone conformation differs in the first 11 residues, particularly between residues 3 and 6. The amino-terminal nine Gla residues are oriented to the interior of the protein and suggest an internal Ca(II) binding pocket. The carboxyl-terminal three Gla residues are exposed to solvent. The majority of hydrophobic residues are required to stabilize a globular core in the carboxyl-terminal three-quarters of the molecule. However, a hydrophobic surface patch in the amino-terminal region may represent a phospholipid binding site in factor IX.
- Center for Hemostasis and Thrombosis Research, New England Medical Center, Boston, Massachusetts, USA.
Organizational Affiliation: