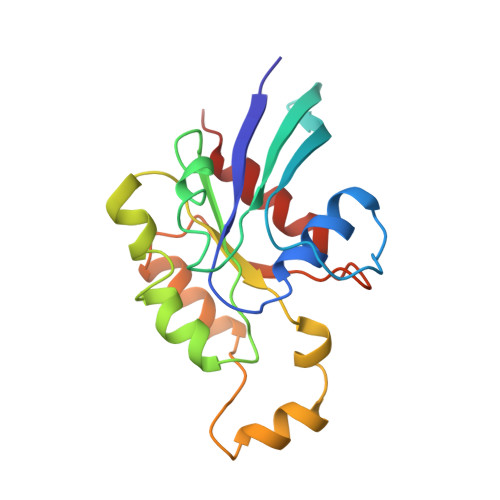
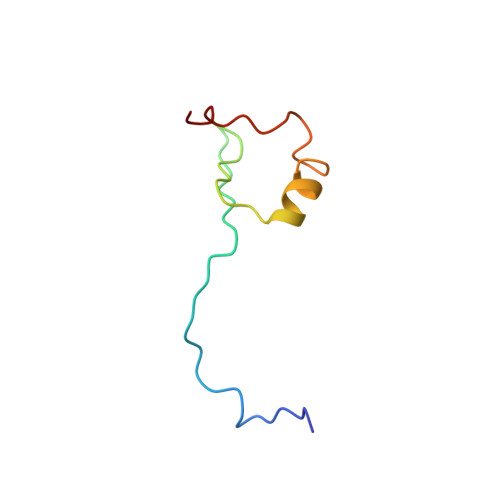
Structure of Cdc42 in complex with the GTPase-binding domain of the 'Wiskott-Aldrich syndrome' protein.
Abdul-Manan, N., Aghazadeh, B., Liu, G.A., Majumdar, A., Ouerfelli, O., Siminovitch, K.A., Rosen, M.K.(1999) Nature 399: 379-383
- PubMed: 10360578
- DOI: https://doi.org/10.1038/20726
- Primary Citation of Related Structures:
1CEE - PubMed Abstract:
The Rho-family GTP-hydrolysing proteins (GTPases), Cdc42, Rac and Rho, act as molecular switches in signalling pathways that regulate cytoskeletal architecture, gene expression and progression of the cell cycle. Cdc42 and Rac transmit many signals through GTP-dependent binding to effector proteins containing a Cdc42/Rac-interactive-binding (CRIB) motif. One such effector, the Wiskott-Aldrich syndrome protein (WASP), is postulated to link activation of Cdc42 directly to the rearrangement of actin. Human mutations in WASP cause severe defects in haematopoletic cell function, leading to clinical symptoms of thrombocytopenia, immunodeficiency and eczema. Here we report the solution structure of a complex between activated Cdc42 and a minimal GTPase-binding domain (GBD) from WASP. An extended amino-terminal GBD peptide that includes the CRIB motif contacts the switch I, beta2 and alpha5 regions of Cdc42. A carboxy-terminal beta-hairpin and alpha-helix pack against switch II. The Phe-X-His-X2-His portion of the CRIB motif and the alpha-helix appear to mediate sensitivity to the nucleotide switch through contacts to residues 36-40 of Cdc42. Discrimination between the Rho-family members is likely to be governed by GBD contacts to the switch I and alpha5 regions of the GTPases. Structural and biochemical data suggest that GBD-sequence divergence outside the CRIB motif may reflect additional regulatory interactions with functional domains that are specific to individual effectors.
- Cellular Biochemistry and Biophysics Program, Memorial Sloan-Kettering Cancer Center, New York, New York 10021, USA.
Organizational Affiliation: