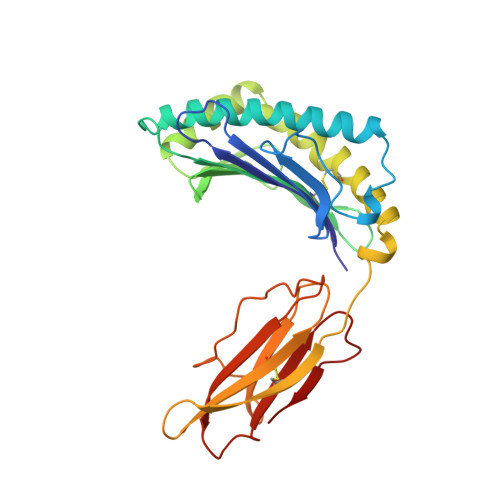
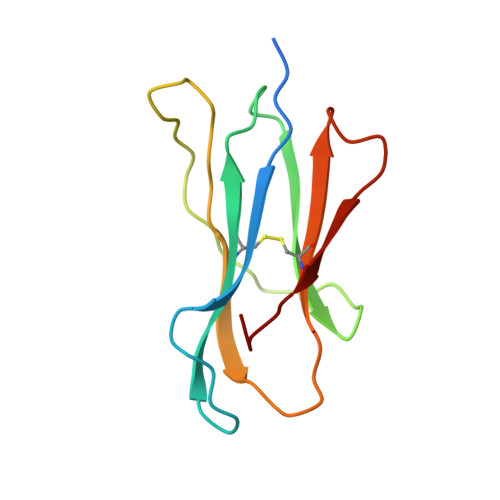
Crystal structures of two H-2Db/glycopeptide complexes suggest a molecular basis for CTL cross-reactivity.
Glithero, A., Tormo, J., Haurum, J.S., Arsequell, G., Valencia, G., Edwards, J., Springer, S., Townsend, A., Pao, Y.L., Wormald, M., Dwek, R.A., Jones, E.Y., Elliott, T.(1999) Immunity 10: 63-74
- PubMed: 10023771
- DOI: https://doi.org/10.1016/s1074-7613(00)80007-2
- Primary Citation of Related Structures:
1CE6, 1QLF - PubMed Abstract:
Two synthetic O-GlcNAc-bearing peptides that elicit H-2Db-restricted glycopeptide-specific cytotoxic T cells (CTL) have been shown to display nonreciprocal patterns of cross-reactivity. Here, we present the crystal structures of the H-2Db glycopeptide complexes to 2.85 A resolution or better. In both cases, the glycan is solvent exposed and available for direct recognition by the T cell receptor (TCR). We have modeled the complex formed between the MHC-glycopeptide complexes and their respective TCRs, showing that a single saccharide residue can be accommodated in the standard TCR-MHC geometry. The models also reveal a possible molecular basis for the observed cross-reactivity patterns of the CTL clones, which appear to be influenced by the length of the CDR3 loop and the nature of the immunizing ligand.
- Nuffield Department of Clinical Medicine, University of Oxford, John Radcliffe Hospital, United Kingdom.
Organizational Affiliation: