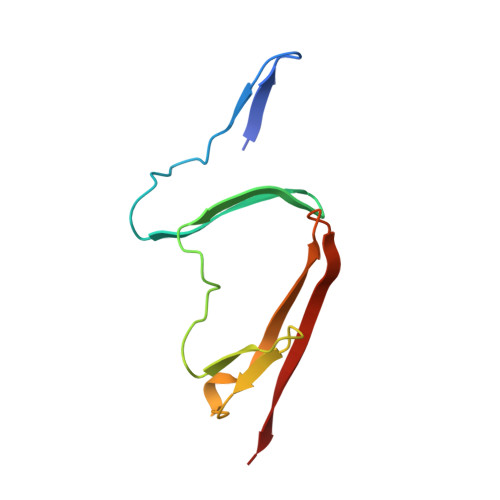
One sequence, two folds: a metastable structure of CD2.
Murray, A.J., Lewis, S.J., Barclay, A.N., Brady, R.L.(1995) Proc Natl Acad Sci U S A 92: 7337-7341
- PubMed: 7638192
- DOI: https://doi.org/10.1073/pnas.92.16.7337
- Primary Citation of Related Structures:
1CDC - PubMed Abstract:
When expressed as part of a glutathione S-transferase fusion protein the NH2-terminal domain of the lymphocyte cell adhesion molecule CD2 is shown to adopt two different folds. The immunoglobulin superfamily structure of the major (85%) monomeric component has previously been determined by both x-ray crystallography and NMR spectroscopy. We now describe the structure of a second, dimeric, form present in about 15% of recombinant CD2 molecules. After denaturation and refolding in the absence of the fusion partner, dimeric CD2 is converted to monomer, illustrating that the dimeric form represents a metastable folded state. The crystal structure of this dimeric form, refined to 2.0-A resolution, reveals two domains with overall similarity to the IgSF fold found in the monomer. However, in the dimer each domain is formed by the intercalation of two polypeptide chains. Hence each domain represents a distinct folding unit that can assemble in two different ways. In the dimer the two domains fold around a hydrophilic interface believed to mimic the cell adhesion interaction at the cell surface, and the formation of dimer can be regulated by mutating single residues at this interface. This unusual misfolded form of the protein, which appears to result from inter- rather than intramolecular interactions being favored by an intermediate structure formed during the folding process, illustrates that evolution of protein oligomers is possible from the sequence for a single protein domain.
- Department of Biochemistry, University of Bristol, United Kingdom.
Organizational Affiliation: