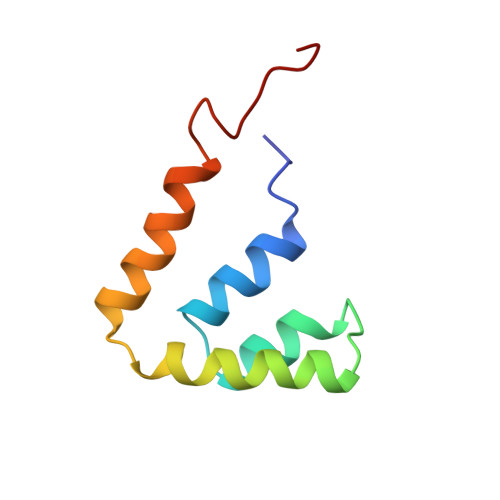
Refined structure of rat Clara cell 17 kDa protein at 3.0 A resolution.
Umland, T.C., Swaminathan, S., Furey, W., Singh, G., Pletcher, J., Sax, M.(1992) J Mol Biology 224: 441-448
- PubMed: 1560460
- DOI: https://doi.org/10.1016/0022-2836(92)91006-b
- Primary Citation of Related Structures:
1CCD - PubMed Abstract:
The rat Clara cell 17 kDa protein (previously referred to as the rat Clara cell 10 kDa protein) has been reported to inhibit phospholipase A2 and papain, and to also bind progesterone. It has been isolated from rat lung lavage fluid and crystallized in the space group P6(5)22. The structure has been determined to 3.0 A resolution using the molecular replacement method. Uteroglobin, whose amino acid sequence is 55.7% identical, was used as the search model. The structure was then refined using restrained least-squares and simulated annealing methods. The R-factor is 22.5%. The protein is a covalently bound dimer. Two disulfide bonds join the monomers together in an antiparallel manner such that the dimer encloses a large internal hydrophobic cavity. The hydrophobic cavity is large enough to serve as the progesterone binding site, but access to the cavity is limited. Each monomer is composed of four alpha-helices. The main-chain structure of the Clara cell protein closely resembles that of uteroglobin, but the nature of many of the exposed side-chains differ. This is true, particularly in a hypervariable region between residues 23 and 36, and in the H1H4 pocket.
- Biocrystallography Laboratory, VA Medical Center, Pittsburgh, PA 15240.
Organizational Affiliation: