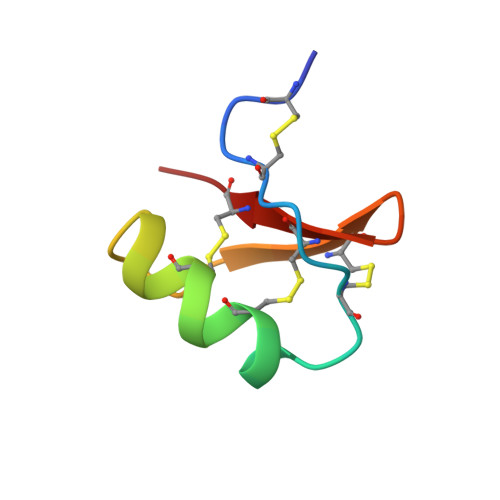
NMR solution structure of butantoxin.
Holaday Jr., S.K., Martin, B.M., Fletcher Jr., P.L., Krishna, N.R.(2000) Arch Biochem Biophys 379: 18-27
- PubMed: 10864437
- DOI: https://doi.org/10.1006/abbi.2000.1858
- Primary Citation of Related Structures:
1C55, 1C56 - PubMed Abstract:
The NMR structure of a new toxin, butantoxin (BuTX), which is present in the venoms of the three Brazilian scorpions Tityus serrulatus, Tityus bahiensis, and Tityus stigmurus, has been investigated. This toxin was shown to reversibly block the Shaker B potassium channels (K(d) approximately 660 nM) and inhibit the proliferation of T-cells and the interleukin-2 production of antigen-stimulated T-helper cells. BuTX is a 40 amino acid basic protein stabilized by the four disulfide bridges: Cys2-Cys5, Cys10-Cys31, Cys16-Cys36, and Cys20-Cys38. The latter three are conserved among all members of the short-chain scorpion toxin family, while the first is unique to BuTX. The three-dimensional structure of BuTX was determined using (1)H-NMR spectroscopy. NOESY, phase sensitive COSY (PH-COSY), and amide hydrogen exchange data were used to generate constraints for molecular modeling calculations. Distance geometry and simulated annealing calculations were performed to generate a family of 49 structures free of constraint violations. The secondary structure of BuTX consists of a short 2(1/2) turn alpha-helix (Glu15-Phe23) and a beta-sheet. The beta-sheet is composed of two well-defined antiparallel strands (Gly29-Met32 and Lys35-Cys38) connected by a type-I' beta-turn (Asn33-Asn34). Residues Cys5-Ala9 form a quasi-third strand of the beta-sheet. The N-terminal C2-C5 disulfide bridge unique to this toxin does not appear to confer stability to the protein.
- Department of Biochemistry and Molecular Genetics, University of Alabama at Birmingham, Birmingham, Alabama, 35294-2041, USA.
Organizational Affiliation: