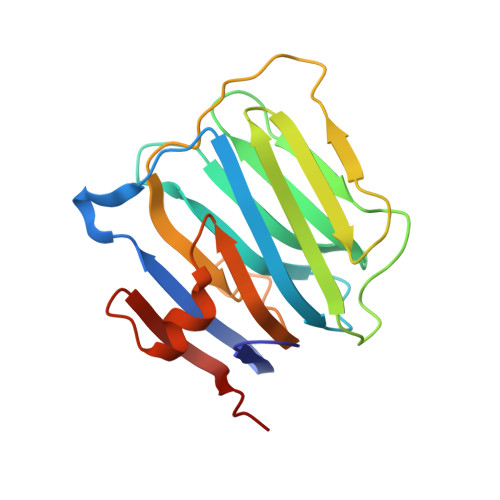
The structure of the ligand-binding domain of neurexin Ibeta: regulation of LNS domain function by alternative splicing.
Rudenko, G., Nguyen, T., Chelliah, Y., Sudhof, T.C., Deisenhofer, J.(1999) Cell 99: 93-101
- PubMed: 10520997
- DOI: https://doi.org/10.1016/s0092-8674(00)80065-3
- Primary Citation of Related Structures:
1C4R - PubMed Abstract:
Neurexins are expressed in hundreds of isoforms on the neuronal cell surface, where they may function as cell recognition molecules. Neurexins contain LNS domains, folding units found in many proteins like the G domain of laminin A, agrin, and slit. The crystal structure of neurexin Ibeta, a single LNS domain, reveals two seven-stranded beta sheets forming a jelly roll fold with unexpected structural similarity to lectins. The LNS domains of neurexin and agrin undergo alternative splicing that modulates their affinity for protein ligands in a neuron-specific manner. These splice sites are localized within loops at one edge of the jelly roll, suggesting a distinct protein interaction surface in LNS domains that is regulated by alternative splicing.
- Howard Hughes Medical Institute, Department of Biochemistry, University of Texas Southwestern Medical Center, Dallas 75235-9050, USA.
Organizational Affiliation: