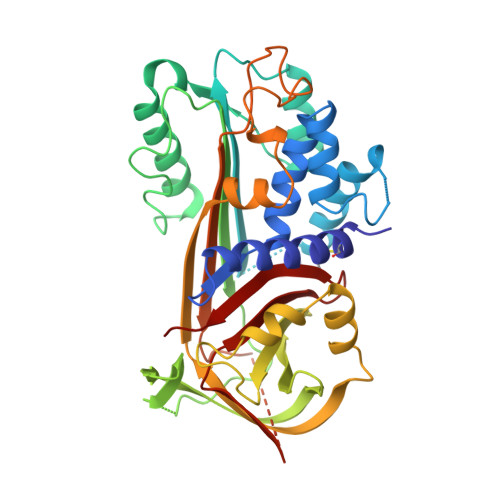
The crystal structure of plasminogen activator inhibitor 2 at 2.0 A resolution: implications for serpin function.
Harrop, S.J., Jankova, L., Coles, M., Jardine, D., Whittaker, J.S., Gould, A.R., Meister, A., King, G.C., Mabbutt, B.C., Curmi, P.M.(1999) Structure 7: 43-54
- PubMed: 10368272
- DOI: https://doi.org/10.1016/s0969-2126(99)80008-2
- Primary Citation of Related Structures:
1BY7 - PubMed Abstract:
Plasminogen activator inhibitor 2 (PAI-2) is a member of the serpin family of protease inhibitors that function via a dramatic structural change from a native, stressed state to a relaxed form. This transition is mediated by a segment of the serpin termed the reactive centre loop (RCL); the RCL is cleaved on interaction with the protease and becomes inserted into betasheet A of the serpin. Major questions remain as to what factors facilitate this transition and how they relate to protease inhibition. The crystal structure of a mutant form of human PAI-2 in the stressed state has been determined at 2.0 A resolution. The RCL is completely disordered in the structure. An examination of polar residues that are highly conserved across all serpins identifies functionally important regions. A buried polar cluster beneath betasheet A (the so-called 'shutter' region) is found to stabilise both the stressed and relaxed forms via a rearrangement of hydrogen bonds. A statistical analysis of interstrand interactions indicated that the shutter region can be used to discriminate between inhibitory and non-inhibitory serpins. This analysis implied that insertion of the RCL into betasheet A up to residue P8 is important for protease inhibition and hence the structure of the complex formed between the serpin and the target protease.
- Initiative in Biomolecular Structure, School of Physics, University of New South Wales, Sydney, NSW 2052, Australia.
Organizational Affiliation: