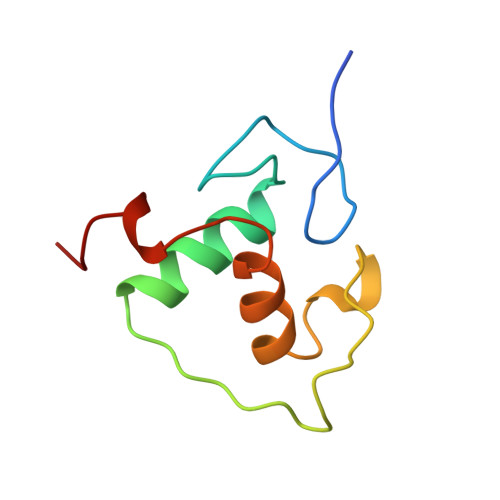
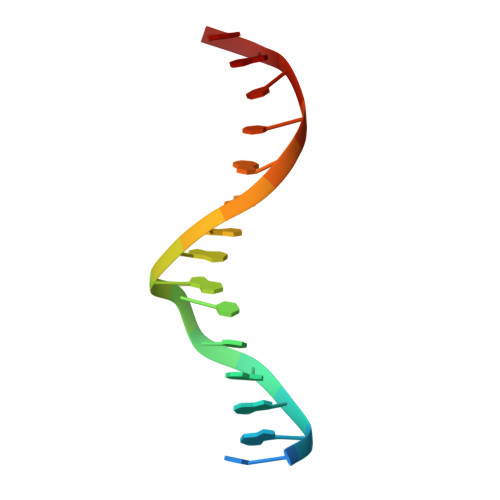
Structural basis of RXR-DNA interactions.
Zhao, Q., Chasse, S.A., Devarakonda, S., Sierk, M.L., Ahvazi, B., Rastinejad, F.(2000) J Mol Biology 296: 509-520
- PubMed: 10669605
- DOI: https://doi.org/10.1006/jmbi.1999.3457
- Primary Citation of Related Structures:
1BY4 - PubMed Abstract:
The 9-cis retinoic acid receptor, RXR, binds DNA effectively as a homodimer or as a heterodimer with other nuclear receptors. The DNA-binding sites for these RXR complexes are direct repeats of a consensus sequence separated by one to five base-pairs of intervening space. Here, we report the 2.1 A crystal structure of the RXR-DNA-binding domain as a homodimer in complex with its idealized direct repeat DNA target. The structure shows how a gene-regulatory site can induce conformational changes in a transcription factor that promote homo-cooperative assembly. Specifically, an alpha-helix in the T-box is disrupted to allow efficient DNA-binding and subunit dimerization. RXR displays a relaxed mode of sequence recognition, interacting with only three base-pairs in each hexameric half-site. The structure illustrates how site selection is achieved in this large eukaryotic transcription factor family through discrete protein-protein interactions and the use of tandem DNA binding sites with characteristic spacings.
- Department of Pharmacology X-ray Crystallography Laboratory, University of Virginia, Charlottesville, VA, 22908, USA.
Organizational Affiliation: