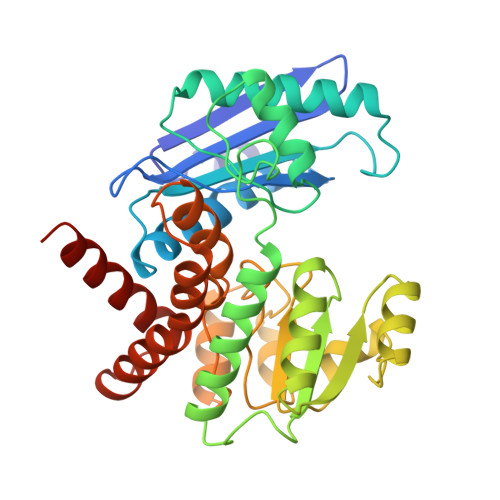
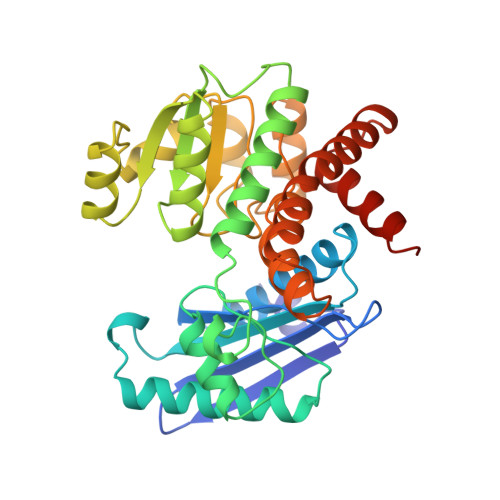
Phenylalanine dehydrogenase from Rhodococcus sp. M4: high-resolution X-ray analyses of inhibitory ternary complexes reveal key features in the oxidative deamination mechanism.
Vanhooke, J.L., Thoden, J.B., Brunhuber, N.M., Blanchard, J.S., Holden, H.M.(1999) Biochemistry 38: 2326-2339
- PubMed: 10029526
- DOI: https://doi.org/10.1021/bi982244q
- Primary Citation of Related Structures:
1BW9, 1BXG - PubMed Abstract:
The molecular structures of recombinant L-phenylalanine dehydrogenase from Rhodococcus sp. M4 in two different inhibitory ternary complexes have been determined by X-ray crystallographic analyses to high resolution. Both structures show that L-phenylalanine dehydrogenase is a homodimeric enzyme with each monomer composed of distinct globular N- and C-terminal domains separated by a deep cleft containing the active site. The N-terminal domain binds the amino acid substrate and contributes to the interactions at the subunit:subunit interface. The C-terminal domain contains a typical Rossmann fold and orients the dinucleotide. The dimer has overall dimensions of approximately 82 A x 75 A x 75 A, with roughly 50 A separating the two active sites. The structures described here, namely the enzyme.NAD+.phenylpyruvate, and enzyme. NAD+.beta-phenylpropionate species, represent the first models for any amino acid dehydrogenase in a ternary complex. By analysis of the active-site interactions in these models, along with the currently available kinetic data, a detailed chemical mechanism has been proposed. This mechanism differs from those proposed to date in that it accounts for the inability of the amino acid dehydrogenases, in general, to function as hydroxy acid dehydrogenases.
- Department of Biochemistry, University of Wisconsin-Madison 53705, USA.
Organizational Affiliation: