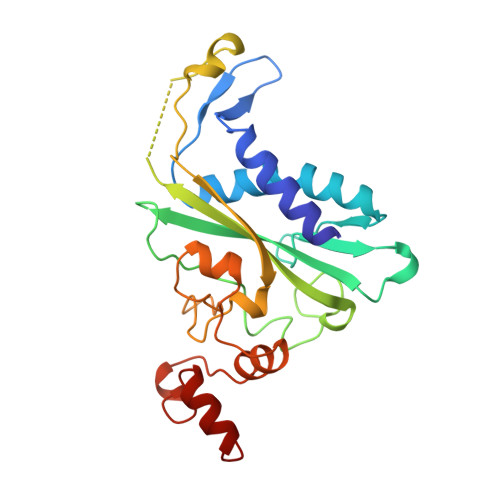
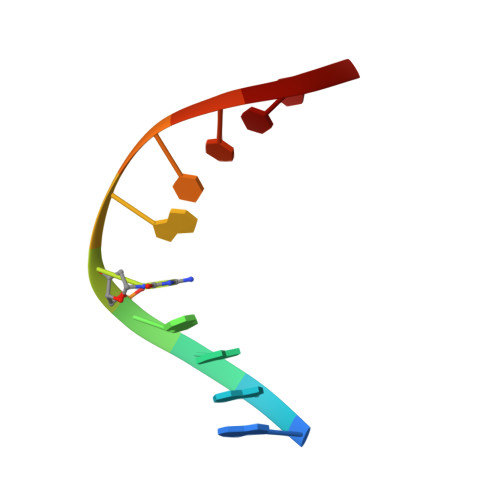
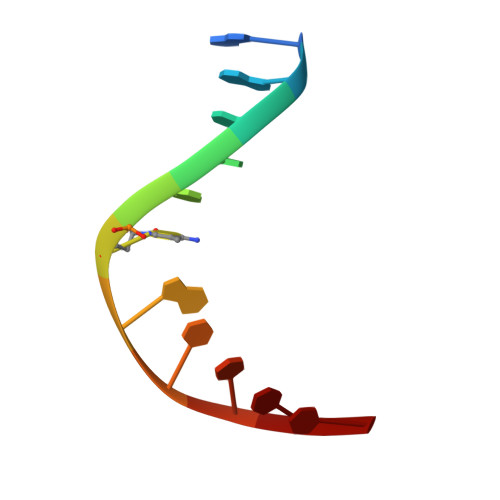
Structural and energetic origins of indirect readout in site-specific DNA cleavage by a restriction endonuclease.
Martin, A.M., Sam, M.D., Reich, N.O., Perona, J.J.(1999) Nat Struct Biol 6: 269-277
- PubMed: 10074946
- DOI: https://doi.org/10.1038/6707
- Primary Citation of Related Structures:
1BSU, 1BUA - PubMed Abstract:
Specific recognition by EcoRV endonuclease of its cognate, sharply bent GATATC site at the center TA step occurs solely via hydrophobic interaction with thymine methyl groups. Mechanistic kinetic analyses of base analog-substituted DNAs at this position reveal that direct readout provides 5 kcal mol(-1) toward specificity, with an additional 6-10 kcal mol(-1) arising from indirect readout. Crystal structures of several base analog complexes show that the major-groove hydrophobic contacts are crucial to forming required divalent metal-binding sites, and that indirect readout operates in part through the sequence-dependent free-energy cost of unstacking the center base-pair step of the DNA.
- Department of Chemistry, University of California at Santa Barbara, 93106-9510, USA.
Organizational Affiliation: