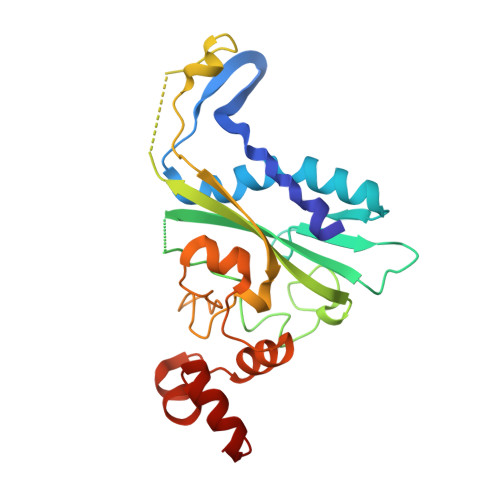
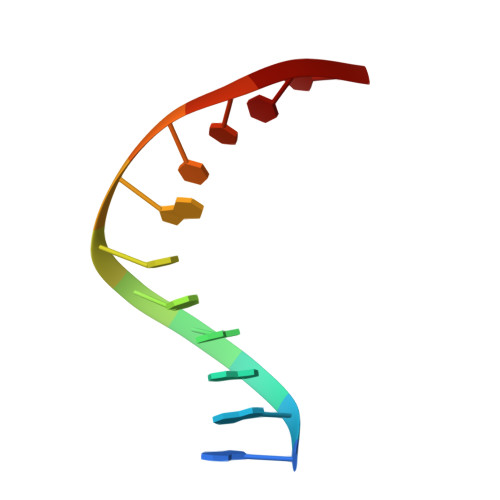
Metal ion-mediated substrate-assisted catalysis in type II restriction endonucleases
Horton, N.C., Newberry, K.J., Perona, J.J.(1998) Proc Natl Acad Sci U S A 95: 13489-13494
- PubMed: 9811827
- DOI: https://doi.org/10.1073/pnas.95.23.13489
- Primary Citation of Related Structures:
1BSS - PubMed Abstract:
The 2.15-A resolution cocrystal structure of EcoRV endonuclease mutant T93A complexed with DNA and Ca2+ ions reveals two divalent metals bound in one of the active sites. One of these metals is ligated through an inner-sphere water molecule to the phosphate group located 3' to the scissile phosphate. A second inner-sphere water on this metal is positioned approximately in-line for attack on the scissile phosphate. This structure corroborates the observation that the pro-SP phosphoryl oxygen on the adjacent 3' phosphate cannot be modified without severe loss of catalytic efficiency. The structural equivalence of key groups, conserved in the active sites of EcoRV, EcoRI, PvuII, and BamHI endonucleases, suggests that ligation of a catalytic divalent metal ion to this phosphate may occur in many type II restriction enzymes. Together with previous cocrystal structures, these data allow construction of a detailed model for the pretransition state configuration in EcoRV. This model features three divalent metal ions per active site and invokes assistance in the bond-making step by a conserved lysine, which stabilizes the attacking hydroxide ion nucleophile.
- Department of Chemistry and Interdepartmental Program in Biochemistry and Molecular Biology, University of California, Santa Barbara, CA 93106-9510, USA.
Organizational Affiliation: